Please be patient as the page loads
|
RPGF4_MOUSE
|
||||||
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RPGF3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.971134 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95398, O95634, Q8WVN0 | Gene names | RAPGEF3, CGEF1, EPAC, EPAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1) (Rap1 guanine-nucleotide-exchange factor directly activated by cAMP). | |||||
|
RPGF3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.965589 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VCC8, Q8BZK9, Q8R1R1 | Gene names | Rapgef3, Epac, Epac1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1). | |||||
|
RPGF4_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.998742 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF4_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
RPGF5_MOUSE
|
||||||
| θ value | 2.03989e-169 (rank : 5) | NC score | 0.935258 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C0Q9, Q8BJJ9, Q8C0R5 | Gene names | Rapgef5, Gfr, Kiaa0277, Mrgef | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
RPGF5_HUMAN
|
||||||
| θ value | 1.42914e-130 (rank : 6) | NC score | 0.926222 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92565, Q8IXU5 | Gene names | RAPGEF5, GFR, KIAA0277, MRGEF | |||
|
Domain Architecture |
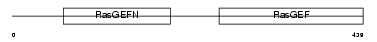
|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (Related to Epac) (Repac) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
RPGF2_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 7) | NC score | 0.697747 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 1.3722e-40 (rank : 8) | NC score | 0.676347 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 2.96777e-27 (rank : 9) | NC score | 0.704876 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 10) | NC score | 0.519281 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 11) | NC score | 0.520910 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 5.99374e-20 (rank : 12) | NC score | 0.520960 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 13) | NC score | 0.505199 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
SOS2_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 14) | NC score | 0.533390 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
GRP3_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 15) | NC score | 0.491338 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
PREX1_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 16) | NC score | 0.200895 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
KAP1_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 17) | NC score | 0.342088 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 18) | NC score | 0.344577 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
RGL1_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 19) | NC score | 0.460391 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZL6, Q9HBY3, Q9HBY4, Q9NZL5, Q9UG43, Q9Y2G6 | Gene names | RGL1, KIAA0959, RGL | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
RGL1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 20) | NC score | 0.428497 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60695 | Gene names | Rgl1, Rgl | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
KGP2_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 21) | NC score | 0.082737 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
CT152_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 22) | NC score | 0.365849 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
KGP2_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 23) | NC score | 0.082534 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
GNDS_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 24) | NC score | 0.433756 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
GP155_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 25) | NC score | 0.218179 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z3F1, Q53SJ3, Q53TA8, Q69YG8, Q86SP9, Q8N261, Q8N639, Q8N8K3, Q96MV6 | Gene names | GPR155, PGR22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR155 (G-protein coupled receptor PGR22). | |||||
|
KAP0_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 26) | NC score | 0.323355 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 27) | NC score | 0.323583 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 28) | NC score | 0.418351 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
CT152_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 29) | NC score | 0.352593 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
RGL2_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 30) | NC score | 0.421390 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61193, Q9QUJ2 | Gene names | Rgl2, Rab2l, Rlf | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
RGL2_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 31) | NC score | 0.415920 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15211, Q9Y3F3 | Gene names | RGL2, RAB2L | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 32) | NC score | 0.075198 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 33) | NC score | 0.075093 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1B_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 34) | NC score | 0.074833 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 35) | NC score | 0.119593 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 36) | NC score | 0.120919 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
KAP2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 37) | NC score | 0.300980 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 38) | NC score | 0.296380 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 39) | NC score | 0.296905 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 40) | NC score | 0.302579 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
SH2D3_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 41) | NC score | 0.130302 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
RGDSR_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 42) | NC score | 0.353475 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZJ4 | Gene names | RGR | |||
|
Domain Architecture |
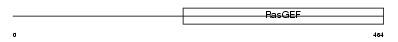
|
|||||
| Description | Ral-GDS-related protein (hRGR). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 43) | NC score | 0.117524 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
DEPD5_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 44) | NC score | 0.086152 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75140, Q5K3V5, Q5THY9, Q5THZ0, Q5THZ1, Q5THZ3, Q68DR1, Q6MZX3, Q6PEZ1, Q9UGV8, Q9UH13 | Gene names | DEPDC5, KIAA0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 45) | NC score | 0.097975 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
PLEK2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 46) | NC score | 0.069084 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |
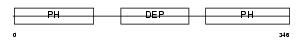
|
|||||
| Description | Pleckstrin-2. | |||||
|
SH2D3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 47) | NC score | 0.121286 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
CNGB3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 48) | NC score | 0.104582 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.073430 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.076315 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.077429 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
DEPD5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.066720 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P61460 | Gene names | Depdc5, Kiaa0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
DVL1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.064517 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |
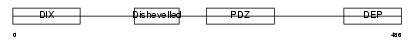
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.049220 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
DVL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.057716 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
CHM1A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.040868 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921W0, Q3TW57 | Gene names | Pcoln3, Chmp1, Chmp1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a). | |||||
|
DVL1L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.059708 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.016583 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.003862 (rank : 91) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.048738 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
PLEK2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.050913 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin-2. | |||||
|
STX1C_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.014783 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61266 | Gene names | STX1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1B2. | |||||
|
STX1C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.014783 (rank : 84) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61264, P32853 | Gene names | Stx1b2, Stx1b | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1B2 (Syntaxin 1B). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.046732 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
DVL3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.056156 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.054880 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.007292 (rank : 88) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.043619 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.043636 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
STX1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.013463 (rank : 85) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16623, O15447, O15448, Q12936, Q9BPZ6 | Gene names | STX1A, STX1 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1A (Neuron-specific antigen HPC-1). | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.017570 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
CHM1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.025664 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HD42, Q14468, Q15779, Q96G31 | Gene names | PCOLN3, CHMP1, CHMP1A, KIAA0047, PRSM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a) (Vacuolar protein sorting 46-1) (Vps46-1) (hVps46-1). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.005781 (rank : 89) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
DYH9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.008029 (rank : 87) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.002555 (rank : 92) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.002246 (rank : 93) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.049312 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.049261 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
STX1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.012424 (rank : 86) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35526 | Gene names | Stx1a | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1A (Neuron-specific antigen HPC-1). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.005459 (rank : 90) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
ARHG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.050537 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.050102 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.069911 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.066648 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.065722 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.065060 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
ECT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.050254 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
HCN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.057258 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.056694 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.056127 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.055988 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.061258 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.064503 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
RPGF4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
RPGF4_HUMAN
|
||||||
| NC score | 0.998742 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF3_HUMAN
|
||||||
| NC score | 0.971134 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95398, O95634, Q8WVN0 | Gene names | RAPGEF3, CGEF1, EPAC, EPAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1) (Rap1 guanine-nucleotide-exchange factor directly activated by cAMP). | |||||
|
RPGF3_MOUSE
|
||||||
| NC score | 0.965589 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VCC8, Q8BZK9, Q8R1R1 | Gene names | Rapgef3, Epac, Epac1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1). | |||||
|
RPGF5_MOUSE
|
||||||
| NC score | 0.935258 (rank : 5) | θ value | 2.03989e-169 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C0Q9, Q8BJJ9, Q8C0R5 | Gene names | Rapgef5, Gfr, Kiaa0277, Mrgef | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
RPGF5_HUMAN
|
||||||
| NC score | 0.926222 (rank : 6) | θ value | 1.42914e-130 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92565, Q8IXU5 | Gene names | RAPGEF5, GFR, KIAA0277, MRGEF | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (Related to Epac) (Repac) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.704876 (rank : 7) | θ value | 2.96777e-27 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
RPGF2_HUMAN
|
||||||
| NC score | 0.697747 (rank : 8) | θ value | 2.76489e-41 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.676347 (rank : 9) | θ value | 1.3722e-40 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
SOS2_HUMAN
|
||||||
| NC score | 0.533390 (rank : 10) | θ value | 1.63225e-17 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.520960 (rank : 11) | θ value | 5.99374e-20 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.520910 (rank : 12) | θ value | 4.58923e-20 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.519281 (rank : 13) | θ value | 3.51386e-20 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.505199 (rank : 14) | θ value | 1.47631e-18 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
GRP3_HUMAN
|
||||||
| NC score | 0.491338 (rank : 15) | θ value | 1.33837e-11 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
RGL1_HUMAN
|
||||||
| NC score | 0.460391 (rank : 16) | θ value | 1.80886e-08 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZL6, Q9HBY3, Q9HBY4, Q9NZL5, Q9UG43, Q9Y2G6 | Gene names | RGL1, KIAA0959, RGL | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.433756 (rank : 17) | θ value | 1.99992e-07 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
RGL1_MOUSE
|
||||||
| NC score | 0.428497 (rank : 18) | θ value | 3.08544e-08 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60695 | Gene names | Rgl1, Rgl | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
RGL2_MOUSE
|
||||||
| NC score | 0.421390 (rank : 19) | θ value | 6.43352e-06 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61193, Q9QUJ2 | Gene names | Rgl2, Rab2l, Rlf | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.418351 (rank : 20) | θ value | 1.29631e-06 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
RGL2_HUMAN
|
||||||
| NC score | 0.415920 (rank : 21) | θ value | 1.09739e-05 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15211, Q9Y3F3 | Gene names | RGL2, RAB2L | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
CT152_HUMAN
|
||||||
| NC score | 0.365849 (rank : 22) | θ value | 6.87365e-08 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
RGDSR_HUMAN
|
||||||
| NC score | 0.353475 (rank : 23) | θ value | 0.0113563 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZJ4 | Gene names | RGR | |||
|
Domain Architecture |

|
|||||
| Description | Ral-GDS-related protein (hRGR). | |||||
|
CT152_MOUSE
|
||||||
| NC score | 0.352593 (rank : 24) | θ value | 1.69304e-06 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.344577 (rank : 25) | θ value | 1.38499e-08 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.342088 (rank : 26) | θ value | 1.06045e-08 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.323583 (rank : 27) | θ value | 9.92553e-07 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.323355 (rank : 28) | θ value | 7.59969e-07 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.302579 (rank : 29) | θ value | 0.00020696 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.300980 (rank : 30) | θ value | 9.29e-05 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.296905 (rank : 31) | θ value | 0.000158464 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.296380 (rank : 32) | θ value | 0.000121331 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
GP155_HUMAN
|
||||||
| NC score | 0.218179 (rank : 33) | θ value | 3.41135e-07 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z3F1, Q53SJ3, Q53TA8, Q69YG8, Q86SP9, Q8N261, Q8N639, Q8N8K3, Q96MV6 | Gene names | GPR155, PGR22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR155 (G-protein coupled receptor PGR22). | |||||
|
PREX1_HUMAN
|
||||||
| NC score | 0.200895 (rank : 34) | θ value | 1.9326e-10 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
SH2D3_MOUSE
|
||||||
| NC score | 0.130302 (rank : 35) | θ value | 0.00869519 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
SH2D3_HUMAN
|
||||||
| NC score | 0.121286 (rank : 36) | θ value | 0.0431538 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.120919 (rank : 37) | θ value | 3.19293e-05 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.119593 (rank : 38) | θ value | 3.19293e-05 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.117524 (rank : 39) | θ value | 0.0193708 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_MOUSE
|
||||||
| NC score | 0.104582 (rank : 40) | θ value | 0.0961366 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.097975 (rank : 41) | θ value | 0.0431538 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
DEPD5_HUMAN
|
||||||
| NC score | 0.086152 (rank : 42) | θ value | 0.0193708 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75140, Q5K3V5, Q5THY9, Q5THZ0, Q5THZ1, Q5THZ3, Q68DR1, Q6MZX3, Q6PEZ1, Q9UGV8, Q9UH13 | Gene names | DEPDC5, KIAA0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
KGP2_HUMAN
|
||||||
| NC score | 0.082737 (rank : 43) | θ value | 5.26297e-08 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KGP2_MOUSE
|
||||||
| NC score | 0.082534 (rank : 44) | θ value | 6.87365e-08 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.077429 (rank : 45) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.076315 (rank : 46) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.075198 (rank : 47) | θ value | 1.43324e-05 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| NC score | 0.075093 (rank : 48) | θ value | 2.44474e-05 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1B_MOUSE
|
||||||
| NC score | 0.074833 (rank : 49) | θ value | 2.44474e-05 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.073430 (rank : 50) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.069911 (rank : 51) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
PLEK2_HUMAN
|
||||||
| NC score | 0.069084 (rank : 52) | θ value | 0.0431538 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
DEPD5_MOUSE
|
||||||
| NC score | 0.066720 (rank : 53) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P61460 | Gene names | Depdc5, Kiaa0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.066648 (rank : 54) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| NC score | 0.065722 (rank : 55) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.065060 (rank : 56) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
DVL1_MOUSE
|
||||||
| NC score | 0.064517 (rank : 57) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.064503 (rank : 58) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.061258 (rank : 59) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
DVL1L_HUMAN
|
||||||
| NC score | 0.059708 (rank : 60) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
DVL1_HUMAN
|
||||||
| NC score | 0.057716 (rank : 61) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.057258 (rank : 62) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.056694 (rank : 63) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.056156 (rank : 64) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.056127 (rank : 65) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.055988 (rank : 66) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.054880 (rank : 67) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
PLEK2_MOUSE
|
||||||
| NC score | 0.050913 (rank : 68) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
ARHG4_HUMAN
|
||||||
| NC score | 0.050537 (rank : 69) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ECT2_HUMAN
|
||||||
| NC score | 0.050254 (rank : 70) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ARHG9_MOUSE
|
||||||
| NC score | 0.050102 (rank : 71) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.049312 (rank : 72) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.049261 (rank : 73) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.049220 (rank : 74) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.048738 (rank : 75) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.046732 (rank : 76) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.043636 (rank : 77) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.043619 (rank : 78) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
CHM1A_MOUSE
|
||||||
| NC score | 0.040868 (rank : 79) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921W0, Q3TW57 | Gene names | Pcoln3, Chmp1, Chmp1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a). | |||||
|
CHM1A_HUMAN
|
||||||
| NC score | 0.025664 (rank : 80) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HD42, Q14468, Q15779, Q96G31 | Gene names | PCOLN3, CHMP1, CHMP1A, KIAA0047, PRSM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a) (Vacuolar protein sorting 46-1) (Vps46-1) (hVps46-1). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.017570 (rank : 81) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.016583 (rank : 82) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
STX1C_HUMAN
|
||||||
| NC score | 0.014783 (rank : 83) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61266 | Gene names | STX1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1B2. | |||||
|
STX1C_MOUSE
|
||||||
| NC score | 0.014783 (rank : 84) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61264, P32853 | Gene names | Stx1b2, Stx1b | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1B2 (Syntaxin 1B). | |||||
|
STX1A_HUMAN
|
||||||
| NC score | 0.013463 (rank : 85) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16623, O15447, O15448, Q12936, Q9BPZ6 | Gene names | STX1A, STX1 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1A (Neuron-specific antigen HPC-1). | |||||
|
STX1A_MOUSE
|
||||||
| NC score | 0.012424 (rank : 86) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35526 | Gene names | Stx1a | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1A (Neuron-specific antigen HPC-1). | |||||
|
DYH9_HUMAN
|
||||||
| NC score | 0.008029 (rank : 87) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.007292 (rank : 88) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.005781 (rank : 89) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.005459 (rank : 90) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.003862 (rank : 91) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.002555 (rank : 92) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.002246 (rank : 93) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||