Please be patient as the page loads
|
RPGF5_MOUSE
|
||||||
| SwissProt Accessions | Q8C0Q9, Q8BJJ9, Q8C0R5 | Gene names | Rapgef5, Gfr, Kiaa0277, Mrgef | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RPGF5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989613 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92565, Q8IXU5 | Gene names | RAPGEF5, GFR, KIAA0277, MRGEF | |||
|
Domain Architecture |
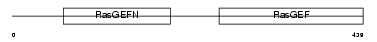
|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (Related to Epac) (Repac) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
RPGF5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8C0Q9, Q8BJJ9, Q8C0R5 | Gene names | Rapgef5, Gfr, Kiaa0277, Mrgef | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
RPGF4_HUMAN
|
||||||
| θ value | 3.36238e-172 (rank : 3) | NC score | 0.936001 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF4_MOUSE
|
||||||
| θ value | 2.03989e-169 (rank : 4) | NC score | 0.935258 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
RPGF3_HUMAN
|
||||||
| θ value | 2.1455e-110 (rank : 5) | NC score | 0.912210 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95398, O95634, Q8WVN0 | Gene names | RAPGEF3, CGEF1, EPAC, EPAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1) (Rap1 guanine-nucleotide-exchange factor directly activated by cAMP). | |||||
|
RPGF3_MOUSE
|
||||||
| θ value | 4.77967e-110 (rank : 6) | NC score | 0.905004 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCC8, Q8BZK9, Q8R1R1 | Gene names | Rapgef3, Epac, Epac1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1). | |||||
|
RPGF2_HUMAN
|
||||||
| θ value | 1.2411e-41 (rank : 7) | NC score | 0.734343 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 1.51715e-39 (rank : 8) | NC score | 0.700217 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 9) | NC score | 0.562974 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 10) | NC score | 0.743255 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 11) | NC score | 0.547499 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 12) | NC score | 0.548563 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 13) | NC score | 0.548143 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS2_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 14) | NC score | 0.562225 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
GRP3_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 15) | NC score | 0.526592 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
RGL1_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 16) | NC score | 0.499415 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZL6, Q9HBY3, Q9HBY4, Q9NZL5, Q9UG43, Q9Y2G6 | Gene names | RGL1, KIAA0959, RGL | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 17) | NC score | 0.456466 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
GNDS_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 18) | NC score | 0.470939 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
RGL1_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 19) | NC score | 0.468419 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60695 | Gene names | Rgl1, Rgl | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
RGL2_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 20) | NC score | 0.459475 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61193, Q9QUJ2 | Gene names | Rgl2, Rab2l, Rlf | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
RGL2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 21) | NC score | 0.451377 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15211, Q9Y3F3 | Gene names | RGL2, RAB2L | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
PREX1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 22) | NC score | 0.179406 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
SH2D3_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 23) | NC score | 0.149813 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
SH2D3_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 24) | NC score | 0.141636 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
RGDSR_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 25) | NC score | 0.385196 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZJ4 | Gene names | RGR | |||
|
Domain Architecture |
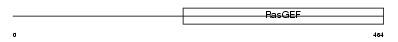
|
|||||
| Description | Ral-GDS-related protein (hRGR). | |||||
|
SAS6_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.051304 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.085251 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.044086 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.027728 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.083362 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.014812 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.019121 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
BLNK_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.039817 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
DEPD5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.058509 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75140, Q5K3V5, Q5THY9, Q5THZ0, Q5THZ1, Q5THZ3, Q68DR1, Q6MZX3, Q6PEZ1, Q9UGV8, Q9UH13 | Gene names | DEPDC5, KIAA0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.026527 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.026485 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.025401 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.034887 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.009220 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
TMCC3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.020887 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULS5, Q8IWB2 | Gene names | TMCC3, KIAA1145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.022665 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.024934 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
RBM19_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.013075 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.023547 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.025268 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.027544 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
DVL1L_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.050531 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |
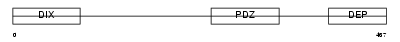
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
GP155_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.141958 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z3F1, Q53SJ3, Q53TA8, Q69YG8, Q86SP9, Q8N261, Q8N639, Q8N8K3, Q96MV6 | Gene names | GPR155, PGR22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR155 (G-protein coupled receptor PGR22). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.026633 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
TPM4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.025019 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
DAAM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.016159 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
DVL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.053662 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |
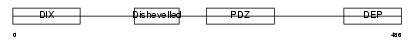
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.019777 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.024421 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.042198 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.042051 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
LRC8D_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.006528 (rank : 95) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L1W4, Q6UWB2, Q9NVW3 | Gene names | LRRC8D, LRRC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
LRC8D_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.006498 (rank : 96) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGR2, Q3UVA9, Q8CI13 | Gene names | Lrrc8d, Lrrc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.022403 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.003282 (rank : 97) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
HD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.012307 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.024106 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.011822 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.013676 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
FBX43_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.012780 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CDI2, Q4G0C8, Q8R0R1 | Gene names | Fbxo43, Emi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 43 (Endogenous meiotic inhibitor 2). | |||||
|
GBP5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.011564 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CFB4, Q8CFA4 | Gene names | Gbp5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 5 (GTP-binding protein 5) (Guanine nucleotide-binding protein 5) (GBP-5) (MuGBP-5). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.021756 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.024341 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.024184 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
PRLR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.008818 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16471, Q16354, Q9BX87 | Gene names | PRLR | |||
|
Domain Architecture |
|
|||||
| Description | Prolactin receptor precursor (PRL-R). | |||||
|
RASF7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.014672 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02833 | Gene names | RASSF7, C11orf13, HRC1 | |||
|
Domain Architecture |
|
|||||
| Description | Ras association domain-containing protein 7 (HRAS1-related cluster protein 1). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.011966 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.011971 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
CFLAR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.009391 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.011366 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
DVL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.047484 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
NEK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | -0.001846 (rank : 99) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35942, O35959 | Gene names | Nek2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek2 (EC 2.7.11.1) (NimA-related protein kinase 2). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.019686 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
UT14C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.009308 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5TAP6, Q5FWG3, Q92555 | Gene names | UTP14C, KIAA0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog C. | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.018097 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.003161 (rank : 98) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.007113 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
REST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.020201 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
VDP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.019455 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
ZN291_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.017518 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
ARHG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.050085 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
BCAR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.055397 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZK2, Q3TNC9, Q3UP10 | Gene names | Bcar3, And34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (p130Cas-binding protein AND-34). | |||||
|
CT152_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.193876 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
CT152_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.163377 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
ECT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.053866 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ECT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.051704 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
KAP0_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.092497 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.092701 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.110777 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.108171 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.083442 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.084684 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.082947 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.083512 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
RPGF5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8C0Q9, Q8BJJ9, Q8C0R5 | Gene names | Rapgef5, Gfr, Kiaa0277, Mrgef | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
RPGF5_HUMAN
|
||||||
| NC score | 0.989613 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92565, Q8IXU5 | Gene names | RAPGEF5, GFR, KIAA0277, MRGEF | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (Related to Epac) (Repac) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
RPGF4_HUMAN
|
||||||
| NC score | 0.936001 (rank : 3) | θ value | 3.36238e-172 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF4_MOUSE
|
||||||
| NC score | 0.935258 (rank : 4) | θ value | 2.03989e-169 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
RPGF3_HUMAN
|
||||||
| NC score | 0.912210 (rank : 5) | θ value | 2.1455e-110 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95398, O95634, Q8WVN0 | Gene names | RAPGEF3, CGEF1, EPAC, EPAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1) (Rap1 guanine-nucleotide-exchange factor directly activated by cAMP). | |||||
|
RPGF3_MOUSE
|
||||||
| NC score | 0.905004 (rank : 6) | θ value | 4.77967e-110 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCC8, Q8BZK9, Q8R1R1 | Gene names | Rapgef3, Epac, Epac1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1). | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.743255 (rank : 7) | θ value | 5.23862e-24 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
RPGF2_HUMAN
|
||||||
| NC score | 0.734343 (rank : 8) | θ value | 1.2411e-41 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.700217 (rank : 9) | θ value | 1.51715e-39 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.562974 (rank : 10) | θ value | 4.01107e-24 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
SOS2_HUMAN
|
||||||
| NC score | 0.562225 (rank : 11) | θ value | 1.52774e-15 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.548563 (rank : 12) | θ value | 9.56915e-18 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.548143 (rank : 13) | θ value | 3.63628e-17 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.547499 (rank : 14) | θ value | 6.40375e-22 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
GRP3_HUMAN
|
||||||
| NC score | 0.526592 (rank : 15) | θ value | 1.2105e-12 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
RGL1_HUMAN
|
||||||
| NC score | 0.499415 (rank : 16) | θ value | 1.80886e-08 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZL6, Q9HBY3, Q9HBY4, Q9NZL5, Q9UG43, Q9Y2G6 | Gene names | RGL1, KIAA0959, RGL | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.470939 (rank : 17) | θ value | 1.99992e-07 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
RGL1_MOUSE
|
||||||
| NC score | 0.468419 (rank : 18) | θ value | 1.99992e-07 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60695 | Gene names | Rgl1, Rgl | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
RGL2_MOUSE
|
||||||
| NC score | 0.459475 (rank : 19) | θ value | 3.77169e-06 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61193, Q9QUJ2 | Gene names | Rgl2, Rab2l, Rlf | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.456466 (rank : 20) | θ value | 8.97725e-08 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
RGL2_HUMAN
|
||||||
| NC score | 0.451377 (rank : 21) | θ value | 5.44631e-05 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15211, Q9Y3F3 | Gene names | RGL2, RAB2L | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
RGDSR_HUMAN
|
||||||
| NC score | 0.385196 (rank : 22) | θ value | 0.0431538 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZJ4 | Gene names | RGR | |||
|
Domain Architecture |

|
|||||
| Description | Ral-GDS-related protein (hRGR). | |||||
|
CT152_HUMAN
|
||||||
| NC score | 0.193876 (rank : 23) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
PREX1_HUMAN
|
||||||
| NC score | 0.179406 (rank : 24) | θ value | 0.000121331 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
CT152_MOUSE
|
||||||
| NC score | 0.163377 (rank : 25) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
SH2D3_MOUSE
|
||||||
| NC score | 0.149813 (rank : 26) | θ value | 0.00035302 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
GP155_HUMAN
|
||||||
| NC score | 0.141958 (rank : 27) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z3F1, Q53SJ3, Q53TA8, Q69YG8, Q86SP9, Q8N261, Q8N639, Q8N8K3, Q96MV6 | Gene names | GPR155, PGR22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR155 (G-protein coupled receptor PGR22). | |||||
|
SH2D3_HUMAN
|
||||||
| NC score | 0.141636 (rank : 28) | θ value | 0.00102713 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.110777 (rank : 29) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.108171 (rank : 30) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.092701 (rank : 31) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.092497 (rank : 32) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.085251 (rank : 33) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.084684 (rank : 34) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.083512 (rank : 35) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.083442 (rank : 36) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.083362 (rank : 37) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.082947 (rank : 38) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
DEPD5_HUMAN
|
||||||
| NC score | 0.058509 (rank : 39) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75140, Q5K3V5, Q5THY9, Q5THZ0, Q5THZ1, Q5THZ3, Q68DR1, Q6MZX3, Q6PEZ1, Q9UGV8, Q9UH13 | Gene names | DEPDC5, KIAA0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
BCAR3_MOUSE
|
||||||
| NC score | 0.055397 (rank : 40) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZK2, Q3TNC9, Q3UP10 | Gene names | Bcar3, And34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (p130Cas-binding protein AND-34). | |||||
|
ECT2_HUMAN
|
||||||
| NC score | 0.053866 (rank : 41) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
DVL1_MOUSE
|
||||||
| NC score | 0.053662 (rank : 42) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
ECT2_MOUSE
|
||||||
| NC score | 0.051704 (rank : 43) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
SAS6_HUMAN
|
||||||
| NC score | 0.051304 (rank : 44) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
DVL1L_HUMAN
|
||||||
| NC score | 0.050531 (rank : 45) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
ARHG4_HUMAN
|
||||||
| NC score | 0.050085 (rank : 46) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
DVL1_HUMAN
|
||||||
| NC score | 0.047484 (rank : 47) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.044086 (rank : 48) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.042198 (rank : 49) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.042051 (rank : 50) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
BLNK_HUMAN
|
||||||
| NC score | 0.039817 (rank : 51) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.034887 (rank : 52) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.027728 (rank : 53) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.027544 (rank : 54) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.026633 (rank : 55) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.026527 (rank : 56) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.026485 (rank : 57) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.025401 (rank : 58) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.025268 (rank : 59) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
TPM4_MOUSE
|
||||||
| NC score | 0.025019 (rank : 60) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.024934 (rank : 61) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.024421 (rank : 62) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.024341 (rank : 63) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.024184 (rank : 64) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.024106 (rank : 65) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.023547 (rank : 66) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.022665 (rank : 67) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.022403 (rank : 68) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.021756 (rank : 69) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
TMCC3_HUMAN
|
||||||
| NC score | 0.020887 (rank : 70) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULS5, Q8IWB2 | Gene names | TMCC3, KIAA1145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.020201 (rank : 71) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.019777 (rank : 72) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.019686 (rank : 73) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
VDP_MOUSE
|
||||||
| NC score | 0.019455 (rank : 74) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.019121 (rank : 75) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.018097 (rank : 76) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
ZN291_HUMAN
|
||||||
| NC score | 0.017518 (rank : 77) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
DAAM2_HUMAN
|
||||||
| NC score | 0.016159 (rank : 78) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.014812 (rank : 79) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
RASF7_HUMAN
|
||||||
| NC score | 0.014672 (rank : 80) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02833 | Gene names | RASSF7, C11orf13, HRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 7 (HRAS1-related cluster protein 1). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.013676 (rank : 81) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RBM19_HUMAN
|
||||||
| NC score | 0.013075 (rank : 82) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
FBX43_MOUSE
|
||||||
| NC score | 0.012780 (rank : 83) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CDI2, Q4G0C8, Q8R0R1 | Gene names | Fbxo43, Emi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 43 (Endogenous meiotic inhibitor 2). | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.012307 (rank : 84) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
APBA2_MOUSE
|
||||||
| NC score | 0.011971 (rank : 85) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.011966 (rank : 86) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.011822 (rank : 87) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
GBP5_MOUSE
|
||||||
| NC score | 0.011564 (rank : 88) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CFB4, Q8CFA4 | Gene names | Gbp5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 5 (GTP-binding protein 5) (Guanine nucleotide-binding protein 5) (GBP-5) (MuGBP-5). | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.011366 (rank : 89) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
CFLAR_MOUSE
|
||||||
| NC score | 0.009391 (rank : 90) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
UT14C_HUMAN
|
||||||
| NC score | 0.009308 (rank : 91) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5TAP6, Q5FWG3, Q92555 | Gene names | UTP14C, KIAA0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog C. | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.009220 (rank : 92) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
PRLR_HUMAN
|
||||||
| NC score | 0.008818 (rank : 93) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16471, Q16354, Q9BX87 | Gene names | PRLR | |||
|
Domain Architecture |

|
|||||
| Description | Prolactin receptor precursor (PRL-R). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.007113 (rank : 94) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
LRC8D_HUMAN
|
||||||
| NC score | 0.006528 (rank : 95) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L1W4, Q6UWB2, Q9NVW3 | Gene names | LRRC8D, LRRC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
LRC8D_MOUSE
|
||||||
| NC score | 0.006498 (rank : 96) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGR2, Q3UVA9, Q8CI13 | Gene names | Lrrc8d, Lrrc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.003282 (rank : 97) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.003161 (rank : 98) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
NEK2_MOUSE
|
||||||
| NC score | -0.001846 (rank : 99) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35942, O35959 | Gene names | Nek2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek2 (EC 2.7.11.1) (NimA-related protein kinase 2). | |||||