Please be patient as the page loads
|
CITE4_MOUSE
|
||||||
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CITE4_MOUSE
|
||||||
| θ value | 5.17823e-64 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
CITE4_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 2) | NC score | 0.904103 (rank : 2) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96RK1 | Gene names | CITED4, MRG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
CITE1_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 3) | NC score | 0.638242 (rank : 3) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99966 | Gene names | CITED1, MSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 1 (Melanocyte-specific protein 1). | |||||
|
CITE2_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 4) | NC score | 0.620262 (rank : 5) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99967, O95426 | Gene names | CITED2, MRG1 | |||
|
Domain Architecture |
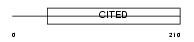
|
|||||
| Description | Cbp/p300-interacting transactivator 2 (MSG-related protein 1) (MRG-1) (P35srj). | |||||
|
CITE2_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 5) | NC score | 0.621869 (rank : 4) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35740, O35741, O35742, O35743, O55198 | Gene names | Cited2, Mrg1, Msg2 | |||
|
Domain Architecture |
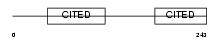
|
|||||
| Description | Cbp/p300-interacting transactivator 2 (MSG-related protein 1) (MRG-1). | |||||
|
CITE1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 6) | NC score | 0.589974 (rank : 6) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97769 | Gene names | Cited1, Msg1 | |||
|
Domain Architecture |
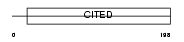
|
|||||
| Description | Cbp/p300-interacting transactivator 1 (Melanocyte-specific protein 1). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.139985 (rank : 7) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.077644 (rank : 53) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.137575 (rank : 8) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.084559 (rank : 38) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.081109 (rank : 45) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
MB3L2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.093823 (rank : 30) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.070541 (rank : 67) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NUP53_MOUSE
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.122038 (rank : 11) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.058623 (rank : 92) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.075474 (rank : 58) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.057558 (rank : 94) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.076653 (rank : 55) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.057163 (rank : 96) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ADA1B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.011255 (rank : 150) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35368 | Gene names | ADRA1B | |||
|
Domain Architecture |
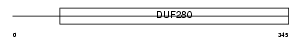
|
|||||
| Description | Alpha-1B adrenergic receptor (Alpha 1B-adrenoceptor) (Alpha 1B- adrenoreceptor). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.076537 (rank : 56) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
TCF19_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.089702 (rank : 32) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
CDX4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.021096 (rank : 141) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.015257 (rank : 143) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.077621 (rank : 54) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.051758 (rank : 122) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.082624 (rank : 41) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
P66A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.081632 (rank : 44) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
NUP53_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.113361 (rank : 12) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
P66A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.081881 (rank : 43) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
IL4RA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.055568 (rank : 105) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |
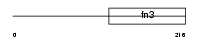
|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
PTSS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.043861 (rank : 127) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVG9 | Gene names | PTDSS2, PSS2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylserine synthase 2 (EC 2.7.8.-) (PtdSer synthase 2) (PSS-2) (Serine-exchange enzyme II). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.012867 (rank : 149) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ACK1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.013151 (rank : 147) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |
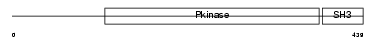
|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.087951 (rank : 36) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
DPOLN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.042845 (rank : 128) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z5Q5, Q4TTW4, Q6ZNF4 | Gene names | POLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase nu (EC 2.7.7.7). | |||||
|
PANK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.030730 (rank : 132) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |
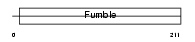
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.095898 (rank : 28) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.068082 (rank : 71) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
FOXK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.022202 (rank : 138) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.027134 (rank : 135) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.037914 (rank : 129) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.030046 (rank : 133) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.021549 (rank : 139) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.030970 (rank : 131) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
AATK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.013767 (rank : 146) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.076459 (rank : 57) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.079170 (rank : 50) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ZIMP7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.122952 (rank : 10) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CBP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.073587 (rank : 64) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.056307 (rank : 98) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
SP5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.006550 (rank : 156) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
ZNHI4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.033654 (rank : 130) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.057405 (rank : 95) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.026997 (rank : 136) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
KSYK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.004463 (rank : 158) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
LHX3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.012925 (rank : 148) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |
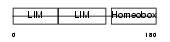
|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
PAX9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.014048 (rank : 145) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-9. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.064305 (rank : 77) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.053737 (rank : 116) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.004374 (rank : 159) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
GAK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.009695 (rank : 153) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
SP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.006248 (rank : 157) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.011026 (rank : 152) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.027744 (rank : 134) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CP007_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.021219 (rank : 140) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
DDFL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.007436 (rank : 154) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.011051 (rank : 151) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
K0552_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.018745 (rank : 142) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60299 | Gene names | KIAA0552 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0552. | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.002705 (rank : 160) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
NADAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.023025 (rank : 137) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
NDE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.006731 (rank : 155) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.014354 (rank : 144) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.075222 (rank : 59) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.054329 (rank : 111) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.070964 (rank : 66) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.052630 (rank : 120) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.063127 (rank : 82) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.084545 (rank : 39) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.055446 (rank : 107) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.060905 (rank : 87) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.050122 (rank : 126) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.111914 (rank : 14) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.108143 (rank : 15) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.051867 (rank : 121) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.088940 (rank : 35) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.090719 (rank : 31) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050281 (rank : 125) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.060895 (rank : 88) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.068164 (rank : 70) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EP300_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.062928 (rank : 83) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.062917 (rank : 84) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
FMN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.055557 (rank : 106) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
FMNL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.057046 (rank : 97) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.078973 (rank : 51) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.074299 (rank : 63) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.079277 (rank : 49) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.101656 (rank : 19) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MISS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.059125 (rank : 91) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.060207 (rank : 89) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.098257 (rank : 24) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.056209 (rank : 102) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.051517 (rank : 124) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053119 (rank : 118) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.106727 (rank : 16) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.103394 (rank : 18) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.053670 (rank : 117) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.111932 (rank : 13) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.134998 (rank : 9) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.099634 (rank : 22) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.089294 (rank : 33) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.051699 (rank : 123) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.089008 (rank : 34) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.099851 (rank : 21) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.058510 (rank : 93) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PYGO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.053768 (rank : 115) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.066262 (rank : 75) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.066666 (rank : 73) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RERE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.064130 (rank : 78) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.056079 (rank : 104) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RSMN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.054518 (rank : 108) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.054518 (rank : 109) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RU1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.059142 (rank : 90) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
RU1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.063594 (rank : 80) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
S23IP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.053818 (rank : 114) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
SC24D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052637 (rank : 119) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
SF01_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.066098 (rank : 76) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SF01_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.053985 (rank : 113) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.097146 (rank : 26) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.098456 (rank : 23) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.063821 (rank : 79) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.062099 (rank : 85) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.074589 (rank : 60) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.078613 (rank : 52) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.098228 (rank : 25) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SMR3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.056144 (rank : 103) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.054091 (rank : 112) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SSBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.056246 (rank : 99) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.056246 (rank : 100) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.066524 (rank : 74) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.054472 (rank : 110) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.080392 (rank : 46) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
T22D4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.056229 (rank : 101) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.100020 (rank : 20) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.067496 (rank : 72) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.070313 (rank : 68) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WASF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.068448 (rank : 69) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.096717 (rank : 27) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.094119 (rank : 29) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.072588 (rank : 65) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.080177 (rank : 47) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.061096 (rank : 86) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
WASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.063476 (rank : 81) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.082207 (rank : 42) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.082903 (rank : 40) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.079765 (rank : 48) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.086077 (rank : 37) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.074420 (rank : 62) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.106508 (rank : 17) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.074466 (rank : 61) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
CITE4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.17823e-64 (rank : 1) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
CITE4_HUMAN
|
||||||
| NC score | 0.904103 (rank : 2) | θ value | 2.95404e-43 (rank : 2) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96RK1 | Gene names | CITED4, MRG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
CITE1_HUMAN
|
||||||
| NC score | 0.638242 (rank : 3) | θ value | 3.29651e-10 (rank : 3) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99966 | Gene names | CITED1, MSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 1 (Melanocyte-specific protein 1). | |||||
|
CITE2_MOUSE
|
||||||
| NC score | 0.621869 (rank : 4) | θ value | 3.29651e-10 (rank : 5) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35740, O35741, O35742, O35743, O55198 | Gene names | Cited2, Mrg1, Msg2 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 2 (MSG-related protein 1) (MRG-1). | |||||
|
CITE2_HUMAN
|
||||||
| NC score | 0.620262 (rank : 5) | θ value | 3.29651e-10 (rank : 4) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99967, O95426 | Gene names | CITED2, MRG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 2 (MSG-related protein 1) (MRG-1) (P35srj). | |||||
|
CITE1_MOUSE
|
||||||
| NC score | 0.589974 (rank : 6) | θ value | 1.69304e-06 (rank : 6) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97769 | Gene names | Cited1, Msg1 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 1 (Melanocyte-specific protein 1). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.139985 (rank : 7) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.137575 (rank : 8) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.134998 (rank : 9) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
ZIMP7_HUMAN
|
||||||
| NC score | 0.122952 (rank : 10) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
NUP53_MOUSE
|
||||||
| NC score | 0.122038 (rank : 11) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
NUP53_HUMAN
|
||||||
| NC score | 0.113361 (rank : 12) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.111932 (rank : 13) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.111914 (rank : 14) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.108143 (rank : 15) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.106727 (rank : 16) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.106508 (rank : 17) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.103394 (rank : 18) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.101656 (rank : 19) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.100020 (rank : 20) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.099851 (rank : 21) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.099634 (rank : 22) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.098456 (rank : 23) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.098257 (rank : 24) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.098228 (rank : 25) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.097146 (rank : 26) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.096717 (rank : 27) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.095898 (rank : 28) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.094119 (rank : 29) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
MB3L2_HUMAN
|
||||||
| NC score | 0.093823 (rank : 30) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.090719 (rank : 31) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.089702 (rank : 32) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.089294 (rank : 33) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.089008 (rank : 34) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.088940 (rank : 35) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.087951 (rank : 36) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.086077 (rank : 37) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.084559 (rank : 38) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.084545 (rank : 39) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.082903 (rank : 40) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.082624 (rank : 41) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.082207 (rank : 42) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
P66A_HUMAN
|
||||||
| NC score | 0.081881 (rank : 43) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
P66A_MOUSE
|
||||||
| NC score | 0.081632 (rank : 44) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.081109 (rank : 45) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.080392 (rank : 46) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.080177 (rank : 47) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.079765 (rank : 48) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.079277 (rank : 49) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.079170 (rank : 50) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.078973 (rank : 51) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.078613 (rank : 52) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.077644 (rank : 53) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.077621 (rank : 54) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.076653 (rank : 55) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.076537 (rank : 56) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.076459 (rank : 57) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.075474 (rank : 58) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.075222 (rank : 59) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.074589 (rank : 60) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.074466 (rank : 61) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.074420 (rank : 62) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.074299 (rank : 63) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.073587 (rank : 64) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.072588 (rank : 65) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.070964 (rank : 66) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.070541 (rank : 67) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.070313 (rank : 68) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.068448 (rank : 69) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.068164 (rank : 70) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.068082 (rank : 71) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.067496 (rank : 72) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.066666 (rank : 73) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.066524 (rank : 74) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.066262 (rank : 75) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.066098 (rank : 76) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.064305 (rank : 77) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.064130 (rank : 78) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.063821 (rank : 79) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
RU1C_MOUSE
|
||||||
| NC score | 0.063594 (rank : 80) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.063476 (rank : 81) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.063127 (rank : 82) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.062928 (rank : 83) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.062917 (rank : 84) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.062099 (rank : 85) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.061096 (rank : 86) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.060905 (rank : 87) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.060895 (rank : 88) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.060207 (rank : 89) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
RU1C_HUMAN
|
||||||
| NC score | 0.059142 (rank : 90) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.059125 (rank : 91) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.058623 (rank : 92) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.058510 (rank : 93) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.057558 (rank : 94) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.057405 (rank : 95) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.057163 (rank : 96) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.057046 (rank : 97) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.056307 (rank : 98) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
SSBP3_HUMAN
|
||||||
| NC score | 0.056246 (rank : 99) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| NC score | 0.056246 (rank : 100) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
T22D4_MOUSE
|
||||||
| NC score | 0.056229 (rank : 101) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056209 (rank : 102) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SMR3B_HUMAN
|
||||||
| NC score | 0.056144 (rank : 103) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.056079 (rank : 104) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
IL4RA_HUMAN
|
||||||
| NC score | 0.055568 (rank : 105) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.055557 (rank : 106) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.055446 (rank : 107) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
RSMN_HUMAN
|
||||||
| NC score | 0.054518 (rank : 108) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| NC score | 0.054518 (rank : 109) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.054472 (rank : 110) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.054329 (rank : 111) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.054091 (rank : 112) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.053985 (rank : 113) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
S23IP_HUMAN
|
||||||
| NC score | 0.053818 (rank : 114) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
PYGO2_HUMAN
|
||||||
| NC score | 0.053768 (rank : 115) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.053737 (rank : 116) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.053670 (rank : 117) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.053119 (rank : 118) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
SC24D_HUMAN
|
||||||
| NC score | 0.052637 (rank : 119) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.052630 (rank : 120) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.051867 (rank : 121) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.051758 (rank : 122) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.051699 (rank : 123) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.051517 (rank : 124) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.050281 (rank : 125) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.050122 (rank : 126) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
PTSS2_HUMAN
|
||||||
| NC score | 0.043861 (rank : 127) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVG9 | Gene names | PTDSS2, PSS2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylserine synthase 2 (EC 2.7.8.-) (PtdSer synthase 2) (PSS-2) (Serine-exchange enzyme II). | |||||
|
DPOLN_HUMAN
|
||||||
| NC score | 0.042845 (rank : 128) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z5Q5, Q4TTW4, Q6ZNF4 | Gene names | POLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase nu (EC 2.7.7.7). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.037914 (rank : 129) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
ZNHI4_MOUSE
|
||||||
| NC score | 0.033654 (rank : 130) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.030970 (rank : 131) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
PANK1_HUMAN
|
||||||
| NC score | 0.030730 (rank : 132) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.030046 (rank : 133) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.027744 (rank : 134) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.027134 (rank : 135) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.026997 (rank : 136) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
NADAP_HUMAN
|
||||||
| NC score | 0.023025 (rank : 137) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
FOXK2_HUMAN
|
||||||
| NC score | 0.022202 (rank : 138) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.021549 (rank : 139) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
CP007_MOUSE
|
||||||
| NC score | 0.021219 (rank : 140) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
CDX4_MOUSE
|
||||||
| NC score | 0.021096 (rank : 141) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
K0552_HUMAN
|
||||||
| NC score | 0.018745 (rank : 142) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60299 | Gene names | KIAA0552 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0552. | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | 0.015257 (rank : 143) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.014354 (rank : 144) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
PAX9_HUMAN
|
||||||
| NC score | 0.014048 (rank : 145) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-9. | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.013767 (rank : 146) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ACK1_MOUSE
|
||||||
| NC score | 0.013151 (rank : 147) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
LHX3_HUMAN
|
||||||
| NC score | 0.012925 (rank : 148) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.012867 (rank : 149) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ADA1B_HUMAN
|
||||||
| NC score | 0.011255 (rank : 150) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35368 | Gene names | ADRA1B | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1B adrenergic receptor (Alpha 1B-adrenoceptor) (Alpha 1B- adrenoreceptor). | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.011051 (rank : 151) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.011026 (rank : 152) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.009695 (rank : 153) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
DDFL1_MOUSE
|
||||||
| NC score | 0.007436 (rank : 154) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
NDE1_MOUSE
|
||||||
| NC score | 0.006731 (rank : 155) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.006550 (rank : 156) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.006248 (rank : 157) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
KSYK_MOUSE
|
||||||
| NC score | 0.004463 (rank : 158) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.004374 (rank : 159) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.002705 (rank : 160) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||