Please be patient as the page loads
|
EPN2_MOUSE
|
||||||
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EPN2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.960883 (rank : 2) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 8.43692e-107 (rank : 3) | NC score | 0.910413 (rank : 5) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 2.54029e-103 (rank : 4) | NC score | 0.912443 (rank : 3) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 4.05562e-101 (rank : 5) | NC score | 0.910827 (rank : 4) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 4.19693e-98 (rank : 6) | NC score | 0.883587 (rank : 6) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN4_HUMAN
|
||||||
| θ value | 1.16164e-39 (rank : 7) | NC score | 0.838251 (rank : 7) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |
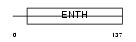
|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| θ value | 1.51715e-39 (rank : 8) | NC score | 0.835871 (rank : 8) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.062080 (rank : 25) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.105617 (rank : 10) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.081838 (rank : 16) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.026707 (rank : 54) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.030033 (rank : 49) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.022221 (rank : 65) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.064689 (rank : 22) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.002728 (rank : 119) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.027419 (rank : 53) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.121112 (rank : 9) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.024480 (rank : 61) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
DSCAM_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.017452 (rank : 76) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60469, O60468 | Gene names | DSCAM | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome cell adhesion molecule precursor (CHD2). | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.098999 (rank : 11) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.027890 (rank : 52) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
MAML1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.035843 (rank : 42) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.060244 (rank : 26) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.085051 (rank : 14) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PIAS3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.037590 (rank : 39) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6X2, Q9UFI3 | Gene names | PIAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.019773 (rank : 71) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.064847 (rank : 21) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DACT1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.030245 (rank : 47) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4A3, Q80VG9, Q8BP49, Q9JK89 | Gene names | Dact1, Thyex3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (MDpr1) (Frodo) (Thymus-expressed novel gene 3 protein). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.055016 (rank : 29) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.067306 (rank : 20) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.034233 (rank : 43) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CJ092_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.038405 (rank : 38) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
NUPL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.033059 (rank : 45) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R332, Q3UJF4, Q8BUA7, Q8BVG7, Q8C0W3 | Gene names | Nupl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p58/p45 (Nucleoporin-like 1). | |||||
|
SOX5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.020475 (rank : 68) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.032106 (rank : 46) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.012940 (rank : 91) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
OBSL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.010685 (rank : 98) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75147, Q96IW3 | Gene names | OBSL1, KIAA0657 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin-like protein 1 precursor. | |||||
|
PIAS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.035883 (rank : 41) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54714, Q80WF8, Q8R598 | Gene names | Pias3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.015854 (rank : 82) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.049108 (rank : 35) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.075342 (rank : 18) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
AP180_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.084969 (rank : 15) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.024744 (rank : 60) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
OBF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.025964 (rank : 56) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64693 | Gene names | Pou2af1, Obf-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain class 2-associating factor 1 (B-cell-specific coactivator OBF-1) (OCT-binding factor 1) (BOB-1) (BOB1) (OCA-B). | |||||
|
TR10C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.026211 (rank : 55) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |
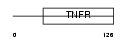
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.011818 (rank : 93) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.033637 (rank : 44) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
T22D4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.017720 (rank : 74) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.037373 (rank : 40) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
ABCAD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.005815 (rank : 116) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
ATF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.023248 (rank : 63) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.023136 (rank : 64) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.020369 (rank : 69) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | -0.000007 (rank : 122) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.016682 (rank : 80) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.006861 (rank : 113) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.024147 (rank : 62) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CUGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.010908 (rank : 97) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92879, Q9NP83, Q9NR06 | Gene names | CUGBP1, BRUNOL2, CUGBP, NAB50 | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (50 kDa Nuclear polyadenylated RNA-binding protein) (EDEN-BP). | |||||
|
CUGB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.010916 (rank : 96) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28659, Q9CXE5, Q9EPJ8, Q9JI37 | Gene names | Cugbp1, Brunol2, Cugbp | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (Deadenylation factor EDEN-BP) (Brain protein F41). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.013298 (rank : 88) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.014473 (rank : 85) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
PDLI5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.006251 (rank : 114) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
RTKN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.016805 (rank : 79) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BST9, Q8WVN1, Q96PT6, Q9HB05 | Gene names | RTKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.017694 (rank : 75) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.030038 (rank : 48) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.053569 (rank : 31) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
TRIP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.014846 (rank : 83) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXN3, Q8CAD5 | Gene names | Trip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 (ASC-1) (Thyroid receptor-interacting protein 4) (Trip-4). | |||||
|
ANR47_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.005745 (rank : 117) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NY19, Q6NZI1, Q6ZQR3, Q8IUV2 | Gene names | ANKRD47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.006161 (rank : 115) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.013640 (rank : 87) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
FA61A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.011649 (rank : 94) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
HDAC6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.006915 (rank : 112) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.006961 (rank : 111) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
K0240_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.020046 (rank : 70) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
LAIR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.011596 (rank : 95) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ISS4, Q6PEZ4 | Gene names | LAIR2, CD306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 2 precursor (LAIR-2) (CD306 antigen). | |||||
|
LYPD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.013851 (rank : 86) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95274, Q9UJ74 | Gene names | LYPD3, C4.4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ly6/PLAUR domain-containing protein 3 precursor (GPI-anchored metastasis-associated protein C4.4A homolog) (Matrigel-induced gene C4 protein) (MIG-C4). | |||||
|
NCTR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.017446 (rank : 77) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95944, Q9H562, Q9H563, Q9H564, Q9UMT1, Q9UMT2 | Gene names | NCR2, LY95 | |||
|
Domain Architecture |
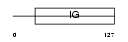
|
|||||
| Description | Natural cytotoxicity triggering receptor 2 precursor (Natural killer cell p44-related protein) (NKp44) (NK-p44) (NK cell-activating receptor) (Lymphocyte antigen 95 homolog) (CD336 antigen). | |||||
|
ONEC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.008457 (rank : 106) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.013068 (rank : 90) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.025803 (rank : 57) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
SIN3B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.008561 (rank : 105) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |
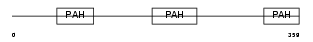
|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.011854 (rank : 92) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
UBQL4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.013143 (rank : 89) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
VPS36_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.018291 (rank : 73) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86VN1, Q3ZCV7, Q5H9S1, Q5VXB6, Q9H8Z5, Q9Y3E3 | Gene names | VPS36, C13orf9, EAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 36 (ELL-associated protein of 45 kDa). | |||||
|
AUXI_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.021285 (rank : 67) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.019172 (rank : 72) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.007429 (rank : 109) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.016495 (rank : 81) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
DGKD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.004368 (rank : 118) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
DYN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.025081 (rank : 59) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.009710 (rank : 102) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.044184 (rank : 36) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LAIR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.010501 (rank : 99) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GTX8 | Gene names | LAIR1, CD305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (hLAIR1) (CD305 antigen). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.001717 (rank : 120) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
LBXCO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.008637 (rank : 104) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.025378 (rank : 58) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MLF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.009937 (rank : 100) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWV4, O70217 | Gene names | Mlf1, Hls7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid leukemia factor 1 (Myelodysplasia-myeloid leukemia factor 1) (Hematopoietic lineage switch 7). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.028133 (rank : 51) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.041713 (rank : 37) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.007182 (rank : 110) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NOC3L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.008024 (rank : 107) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.029083 (rank : 50) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PHF12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.009862 (rank : 101) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PRCC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.021466 (rank : 66) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
PS1C2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.085760 (rank : 13) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.014724 (rank : 84) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.009527 (rank : 103) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
TMC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.007983 (rank : 108) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
VPS36_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.016952 (rank : 78) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91XD6, Q9CVA3 | Gene names | Vps36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 36. | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.001597 (rank : 121) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
AP180_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.063234 (rank : 24) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.090781 (rank : 12) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.070437 (rank : 19) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
MAML1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.050337 (rank : 33) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
MISS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.064263 (rank : 23) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.053679 (rank : 30) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.077022 (rank : 17) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.050245 (rank : 34) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.052452 (rank : 32) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.059321 (rank : 27) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TCF19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.056708 (rank : 28) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
EPN2_HUMAN
|
||||||
| NC score | 0.960883 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.912443 (rank : 3) | θ value | 2.54029e-103 (rank : 4) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.910827 (rank : 4) | θ value | 4.05562e-101 (rank : 5) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.910413 (rank : 5) | θ value | 8.43692e-107 (rank : 3) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.883587 (rank : 6) | θ value | 4.19693e-98 (rank : 6) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN4_HUMAN
|
||||||
| NC score | 0.838251 (rank : 7) | θ value | 1.16164e-39 (rank : 7) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| NC score | 0.835871 (rank : 8) | θ value | 1.51715e-39 (rank : 8) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.121112 (rank : 9) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.105617 (rank : 10) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.098999 (rank : 11) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.090781 (rank : 12) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
PS1C2_MOUSE
|
||||||
| NC score | 0.085760 (rank : 13) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.085051 (rank : 14) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
AP180_MOUSE
|
||||||
| NC score | 0.084969 (rank : 15) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.081838 (rank : 16) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.077022 (rank : 17) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.075342 (rank : 18) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.070437 (rank : 19) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.067306 (rank : 20) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.064847 (rank : 21) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.064689 (rank : 22) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.064263 (rank : 23) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.063234 (rank : 24) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.062080 (rank : 25) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.060244 (rank : 26) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.059321 (rank : 27) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.056708 (rank : 28) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.055016 (rank : 29) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.053679 (rank : 30) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.053569 (rank : 31) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.052452 (rank : 32) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MAML1_MOUSE
|
||||||
| NC score | 0.050337 (rank : 33) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.050245 (rank : 34) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.049108 (rank : 35) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.044184 (rank : 36) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.041713 (rank : 37) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CJ092_HUMAN
|
||||||
| NC score | 0.038405 (rank : 38) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
PIAS3_HUMAN
|
||||||
| NC score | 0.037590 (rank : 39) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6X2, Q9UFI3 | Gene names | PIAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.037373 (rank : 40) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
PIAS3_MOUSE
|
||||||
| NC score | 0.035883 (rank : 41) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54714, Q80WF8, Q8R598 | Gene names | Pias3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
MAML1_HUMAN
|
||||||
| NC score | 0.035843 (rank : 42) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.034233 (rank : 43) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.033637 (rank : 44) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NUPL1_MOUSE
|
||||||
| NC score | 0.033059 (rank : 45) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R332, Q3UJF4, Q8BUA7, Q8BVG7, Q8C0W3 | Gene names | Nupl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p58/p45 (Nucleoporin-like 1). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.032106 (rank : 46) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
DACT1_MOUSE
|
||||||
| NC score | 0.030245 (rank : 47) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4A3, Q80VG9, Q8BP49, Q9JK89 | Gene names | Dact1, Thyex3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (MDpr1) (Frodo) (Thymus-expressed novel gene 3 protein). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.030038 (rank : 48) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.030033 (rank : 49) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.029083 (rank : 50) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.028133 (rank : 51) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.027890 (rank : 52) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.027419 (rank : 53) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.026707 (rank : 54) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
TR10C_HUMAN
|
||||||
| NC score | 0.026211 (rank : 55) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
OBF1_MOUSE
|
||||||
| NC score | 0.025964 (rank : 56) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64693 | Gene names | Pou2af1, Obf-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain class 2-associating factor 1 (B-cell-specific coactivator OBF-1) (OCT-binding factor 1) (BOB-1) (BOB1) (OCA-B). | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.025803 (rank : 57) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.025378 (rank : 58) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
DYN1_HUMAN
|
||||||
| NC score | 0.025081 (rank : 59) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.024744 (rank : 60) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.024480 (rank : 61) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.024147 (rank : 62) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.023248 (rank : 63) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.023136 (rank : 64) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.022221 (rank : 65) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
PRCC_HUMAN
|
||||||
| NC score | 0.021466 (rank : 66) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.021285 (rank : 67) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
SOX5_MOUSE
|
||||||
| NC score | 0.020475 (rank : 68) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.020369 (rank : 69) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
K0240_MOUSE
|
||||||
| NC score | 0.020046 (rank : 70) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.019773 (rank : 71) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.019172 (rank : 72) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
VPS36_HUMAN
|
||||||
| NC score | 0.018291 (rank : 73) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86VN1, Q3ZCV7, Q5H9S1, Q5VXB6, Q9H8Z5, Q9Y3E3 | Gene names | VPS36, C13orf9, EAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 36 (ELL-associated protein of 45 kDa). | |||||
|
T22D4_MOUSE
|
||||||
| NC score | 0.017720 (rank : 74) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.017694 (rank : 75) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
DSCAM_HUMAN
|
||||||
| NC score | 0.017452 (rank : 76) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60469, O60468 | Gene names | DSCAM | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome cell adhesion molecule precursor (CHD2). | |||||
|
NCTR2_HUMAN
|
||||||
| NC score | 0.017446 (rank : 77) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95944, Q9H562, Q9H563, Q9H564, Q9UMT1, Q9UMT2 | Gene names | NCR2, LY95 | |||
|
Domain Architecture |

|
|||||
| Description | Natural cytotoxicity triggering receptor 2 precursor (Natural killer cell p44-related protein) (NKp44) (NK-p44) (NK cell-activating receptor) (Lymphocyte antigen 95 homolog) (CD336 antigen). | |||||
|
VPS36_MOUSE
|
||||||
| NC score | 0.016952 (rank : 78) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91XD6, Q9CVA3 | Gene names | Vps36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 36. | |||||
|
RTKN_HUMAN
|
||||||
| NC score | 0.016805 (rank : 79) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BST9, Q8WVN1, Q96PT6, Q9HB05 | Gene names | RTKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.016682 (rank : 80) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.016495 (rank : 81) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.015854 (rank : 82) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
TRIP4_MOUSE
|
||||||
| NC score | 0.014846 (rank : 83) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXN3, Q8CAD5 | Gene names | Trip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 (ASC-1) (Thyroid receptor-interacting protein 4) (Trip-4). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.014724 (rank : 84) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.014473 (rank : 85) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
LYPD3_HUMAN
|
||||||
| NC score | 0.013851 (rank : 86) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95274, Q9UJ74 | Gene names | LYPD3, C4.4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ly6/PLAUR domain-containing protein 3 precursor (GPI-anchored metastasis-associated protein C4.4A homolog) (Matrigel-induced gene C4 protein) (MIG-C4). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.013640 (rank : 87) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.013298 (rank : 88) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
UBQL4_MOUSE
|
||||||
| NC score | 0.013143 (rank : 89) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.013068 (rank : 90) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.012940 (rank : 91) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.011854 (rank : 92) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.011818 (rank : 93) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
FA61A_MOUSE
|
||||||
| NC score | 0.011649 (rank : 94) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
LAIR2_HUMAN
|
||||||
| NC score | 0.011596 (rank : 95) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ISS4, Q6PEZ4 | Gene names | LAIR2, CD306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 2 precursor (LAIR-2) (CD306 antigen). | |||||
|
CUGB1_MOUSE
|
||||||
| NC score | 0.010916 (rank : 96) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28659, Q9CXE5, Q9EPJ8, Q9JI37 | Gene names | Cugbp1, Brunol2, Cugbp | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (Deadenylation factor EDEN-BP) (Brain protein F41). | |||||
|
CUGB1_HUMAN
|
||||||
| NC score | 0.010908 (rank : 97) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92879, Q9NP83, Q9NR06 | Gene names | CUGBP1, BRUNOL2, CUGBP, NAB50 | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (50 kDa Nuclear polyadenylated RNA-binding protein) (EDEN-BP). | |||||
|
OBSL1_HUMAN
|
||||||
| NC score | 0.010685 (rank : 98) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75147, Q96IW3 | Gene names | OBSL1, KIAA0657 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin-like protein 1 precursor. | |||||
|
LAIR1_HUMAN
|
||||||
| NC score | 0.010501 (rank : 99) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GTX8 | Gene names | LAIR1, CD305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (hLAIR1) (CD305 antigen). | |||||
|
MLF1_MOUSE
|
||||||
| NC score | 0.009937 (rank : 100) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWV4, O70217 | Gene names | Mlf1, Hls7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid leukemia factor 1 (Myelodysplasia-myeloid leukemia factor 1) (Hematopoietic lineage switch 7). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.009862 (rank : 101) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.009710 (rank : 102) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.009527 (rank : 103) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
LBXCO_HUMAN
|
||||||
| NC score | 0.008637 (rank : 104) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
SIN3B_HUMAN
|
||||||
| NC score | 0.008561 (rank : 105) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
ONEC2_MOUSE
|
||||||
| NC score | 0.008457 (rank : 106) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
NOC3L_MOUSE
|
||||||
| NC score | 0.008024 (rank : 107) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
TMC2_HUMAN
|
||||||
| NC score | 0.007983 (rank : 108) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.007429 (rank : 109) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.007182 (rank : 110) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.006961 (rank : 111) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
HDAC6_MOUSE
|
||||||
| NC score | 0.006915 (rank : 112) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.006861 (rank : 113) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
PDLI5_HUMAN
|
||||||
| NC score | 0.006251 (rank : 114) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.006161 (rank : 115) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
ABCAD_MOUSE
|
||||||
| NC score | 0.005815 (rank : 116) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
ANR47_HUMAN
|
||||||
| NC score | 0.005745 (rank : 117) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NY19, Q6NZI1, Q6ZQR3, Q8IUV2 | Gene names | ANKRD47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
DGKD_HUMAN
|
||||||
| NC score | 0.004368 (rank : 118) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.002728 (rank : 119) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.001717 (rank : 120) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.001597 (rank : 121) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | -0.000007 (rank : 122) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||