Please be patient as the page loads
|
UBQL4_MOUSE
|
||||||
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBQL4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984518 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NRR5, Q9BR98, Q9UHX4 | Gene names | UBQLN4, C1orf6, UBIN | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
UBQL4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
UBQL1_MOUSE
|
||||||
| θ value | 9.47563e-167 (rank : 3) | NC score | 0.979512 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R317, Q80V10, Q8C7T4, Q8C835, Q8K141, Q91VI8, Q9D0Z0, Q9QZM1 | Gene names | Ubqln1, Plic1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-1 (Protein linking IAP with cytoskeleton 1) (PLIC-1). | |||||
|
UBQL1_HUMAN
|
||||||
| θ value | 4.42207e-148 (rank : 4) | NC score | 0.982183 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UMX0, Q8IXS9, Q8N2Q3, Q9H0T8, Q9H3R4, Q9HAZ5 | Gene names | UBQLN1, DA41, PLIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-1 (Protein linking IAP with cytoskeleton 1) (PLIC-1) (hPLIC- 1). | |||||
|
UBQL2_MOUSE
|
||||||
| θ value | 2.689e-137 (rank : 5) | NC score | 0.972878 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZM0, Q7TSJ8, Q8VDH9 | Gene names | Ubqln2, Plic2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
UBQL2_HUMAN
|
||||||
| θ value | 1.58745e-113 (rank : 6) | NC score | 0.969044 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHD9, O94798, Q9H3W6, Q9HAZ4 | Gene names | UBQLN2, PLIC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (hPLIC- 2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
UBQL3_HUMAN
|
||||||
| θ value | 2.3998e-69 (rank : 7) | NC score | 0.910973 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
UBQL3_MOUSE
|
||||||
| θ value | 3.37202e-47 (rank : 8) | NC score | 0.892694 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C5U9 | Gene names | Ubqln3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
UBL7_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 9) | NC score | 0.371139 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96S82, Q96I03 | Gene names | UBL7, BMSCUBP | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-like protein 7 (Ubiquitin-like protein SB132) (Bone marrow stromal cell ubiquitin-like protein) (BMSC-UbP). | |||||
|
UBL7_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 10) | NC score | 0.371571 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 11) | NC score | 0.234265 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 12) | NC score | 0.225453 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
UBIQ_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 13) | NC score | 0.313814 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62988, P02248, P02249, P02250, Q29120, Q6LBL4, Q6LDU5, Q91887, Q91888, Q9BWD6, Q9BX98, Q9UEF2, Q9UEG1, Q9UEK8, Q9UPK7 | Gene names | RPS27A, UBA80, UBCEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin. | |||||
|
UBIQ_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.313814 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62991, P02248, P02249, P02250, Q29120, Q62317, Q64223, Q8VCH1, Q91887, Q91888, Q9CXY4, Q9CZM0, Q9D1R5, Q9D8D9, Q9ET23, Q9ET24, Q9Z0H9 | Gene names | Rps27a, Uba80, Ubcep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin. | |||||
|
UBD_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 15) | NC score | 0.292203 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15205, Q5SUK2, Q96EC7 | Gene names | UBD, FAT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin D (Ubiquitin-like protein FAT10) (Diubiquitin). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.048311 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
PRKN2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.182590 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.045753 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
UBL4A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.230409 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P11441 | Gene names | UBL4A, DXS254E, GDX, UBL4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 4A (Ubiquitin-like protein GDX). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.069035 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
PRKN2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.195499 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WVS6, Q9ES22, Q9ES23 | Gene names | Park2, Prkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.038604 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
TMUB2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.149588 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3V209, Q8K0X0, Q91YQ0, Q9D8C5 | Gene names | Tmub2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
UBD_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.275241 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63072, Q9WV10 | Gene names | Ubd, Fat10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin D (Ubiquitin-like protein FAT10) (Diubiquitin). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.033946 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SYRC_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.071136 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54136, Q9BWA1 | Gene names | RARS | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA synthetase, cytoplasmic (EC 6.1.1.19) (Arginine--tRNA ligase) (ArgRS). | |||||
|
NEDD8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.238571 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15843, Q6LES6 | Gene names | NEDD8 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 precursor (Ubiquitin-like protein Nedd8) (Neddylin). | |||||
|
NEDD8_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.238300 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29595 | Gene names | Nedd8, Nedd-8 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 precursor (Ubiquitin-like protein Nedd8) (Neddylin) (Neural precursor cell expressed developmentally down-regulated protein 8). | |||||
|
RD23A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.148158 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P54725 | Gene names | RAD23A | |||
|
Domain Architecture |
|
|||||
| Description | UV excision repair protein RAD23 homolog A (hHR23A). | |||||
|
UCRP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.213158 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64339, Q8K0H3 | Gene names | Isg15, G1p2, Ucrp | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin cross-reactive protein precursor (Interferon-stimulated protein 15) (IP17). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.008194 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
SYRC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.062194 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0I9, Q8VDW1 | Gene names | Rars | |||
|
Domain Architecture |
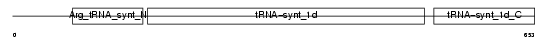
|
|||||
| Description | Arginyl-tRNA synthetase, cytoplasmic (EC 6.1.1.19) (Arginine--tRNA ligase) (ArgRS). | |||||
|
DCC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.006841 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
RNF31_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.078996 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.015789 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.020847 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
UBL4A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.169598 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P21126 | Gene names | Ubl4a, Gdx, Ubl4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 4A (Ubiquitin-like protein GDX). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.029125 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
RNF31_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.068078 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
SATB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.035119 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01826 | Gene names | SATB1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
SATB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.035475 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60611 | Gene names | Satb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.024909 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
KLF11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.001408 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14901 | Gene names | KLF11, FKLF, TIEG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 11 (Transforming growth factor-beta-inducible early growth response protein 2) (TGFB-inducible early growth response protein 2) (TIEG-2). | |||||
|
NUP62_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.020561 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
RD23A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.121881 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54726 | Gene names | Rad23a, Mhr23a | |||
|
Domain Architecture |
|
|||||
| Description | UV excision repair protein RAD23 homolog A (mHR23A). | |||||
|
THSD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.015822 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
YETS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.020875 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
ADRM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.033646 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.024693 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
IRAK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.003149 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |
|
|||||
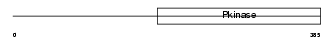
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
JHD2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.014155 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.095119 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
TMUB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.132310 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q71RG4, Q8NDI2, Q9BPZ5, Q9HAG3 | Gene names | TMUB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.044977 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.026777 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.014180 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
RD23B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.099437 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P54727, Q8WUB0 | Gene names | RAD23B | |||
|
Domain Architecture |
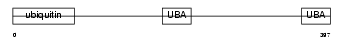
|
|||||
| Description | UV excision repair protein RAD23 homolog B (hHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.006219 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
ADRM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.029742 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |
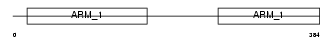
|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
DGCR8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.021034 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.009446 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HXD12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.009091 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23812 | Gene names | Hoxd12, Hox-4.7, Hoxd-12 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4.7) (Hox-5.6). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.016705 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.019022 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RINT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.022729 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ36, Q3UXS1, Q6P1I2, Q8C193 | Gene names | Rint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD50-interacting protein 1 (RAD50 interactor 1) (Protein RINT-1). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.017127 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
TDRD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.014315 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.009508 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.006432 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.021452 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.012979 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.019551 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
AMZ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.015162 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVF9, Q811F9, Q8BMM5 | Gene names | Amz1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Archaemetzincin-1 (EC 3.-.-.-) (Archeobacterial metalloproteinase-like protein 1). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.013962 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.035290 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.013143 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
FOXP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.020611 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
ISL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.001967 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96A47 | Gene names | ISL2 | |||
|
Domain Architecture |
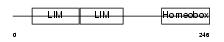
|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
ITIH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.006363 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
KLF17_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.001976 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
LRC8D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.001852 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L1W4, Q6UWB2, Q9NVW3 | Gene names | LRRC8D, LRRC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.015639 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.016711 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
RAD9A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.010615 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0F6, Q8QZZ3 | Gene names | Rad9a, Rad9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint control protein RAD9A (EC 3.1.11.2) (DNA repair exonuclease rad9 homolog A) (Rad9-like protein) (mRAD9). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.003773 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.010442 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.010148 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.003660 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
DPEP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.006029 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DA79 | Gene names | Dpep3, Mbd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidase 3 precursor (EC 3.4.13.19) (Membrane-bound dipeptidase 3) (MBD-3). | |||||
|
DPOE4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.011292 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQ36, Q3TJ13 | Gene names | Pole4 | |||
|
Domain Architecture |
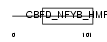
|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.002839 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
FATH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | -0.000475 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
K1849_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.008032 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
MBRL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.009922 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q4ZIN3, O60392, Q8NF79, Q96H30 | Gene names | C19orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membralin. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.018318 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.008591 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.001771 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.010471 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.004379 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
UBIM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.075058 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35544, Q9H5V4 | Gene names | FAU | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein FUBI. | |||||
|
UBIM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.070983 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35545, Q9JJ24 | Gene names | Fau | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein FUBI (Monoclonal nonspecific suppressor factor beta) (MNSF-beta). | |||||
|
UCRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.059366 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05161, Q7Z2G2, Q96GF0 | Gene names | ISG15, G1P2, UCRP | |||
|
Domain Architecture |
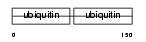
|
|||||
| Description | Interferon-induced 17 kDa protein precursor [Contains: Ubiquitin cross-reactive protein (hUCRP) (Interferon-induced 15 kDa protein)]. | |||||
|
UBQL4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
UBQL4_HUMAN
|
||||||
| NC score | 0.984518 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NRR5, Q9BR98, Q9UHX4 | Gene names | UBQLN4, C1orf6, UBIN | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
UBQL1_HUMAN
|
||||||
| NC score | 0.982183 (rank : 3) | θ value | 4.42207e-148 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UMX0, Q8IXS9, Q8N2Q3, Q9H0T8, Q9H3R4, Q9HAZ5 | Gene names | UBQLN1, DA41, PLIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-1 (Protein linking IAP with cytoskeleton 1) (PLIC-1) (hPLIC- 1). | |||||
|
UBQL1_MOUSE
|
||||||
| NC score | 0.979512 (rank : 4) | θ value | 9.47563e-167 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R317, Q80V10, Q8C7T4, Q8C835, Q8K141, Q91VI8, Q9D0Z0, Q9QZM1 | Gene names | Ubqln1, Plic1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-1 (Protein linking IAP with cytoskeleton 1) (PLIC-1). | |||||
|
UBQL2_MOUSE
|
||||||
| NC score | 0.972878 (rank : 5) | θ value | 2.689e-137 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZM0, Q7TSJ8, Q8VDH9 | Gene names | Ubqln2, Plic2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
UBQL2_HUMAN
|
||||||
| NC score | 0.969044 (rank : 6) | θ value | 1.58745e-113 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHD9, O94798, Q9H3W6, Q9HAZ4 | Gene names | UBQLN2, PLIC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (hPLIC- 2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
UBQL3_HUMAN
|
||||||
| NC score | 0.910973 (rank : 7) | θ value | 2.3998e-69 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
UBQL3_MOUSE
|
||||||
| NC score | 0.892694 (rank : 8) | θ value | 3.37202e-47 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C5U9 | Gene names | Ubqln3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
UBL7_MOUSE
|
||||||
| NC score | 0.371571 (rank : 9) | θ value | 4.1701e-05 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
UBL7_HUMAN
|
||||||
| NC score | 0.371139 (rank : 10) | θ value | 4.1701e-05 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96S82, Q96I03 | Gene names | UBL7, BMSCUBP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 7 (Ubiquitin-like protein SB132) (Bone marrow stromal cell ubiquitin-like protein) (BMSC-UbP). | |||||
|
UBIQ_HUMAN
|
||||||
| NC score | 0.313814 (rank : 11) | θ value | 0.00509761 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62988, P02248, P02249, P02250, Q29120, Q6LBL4, Q6LDU5, Q91887, Q91888, Q9BWD6, Q9BX98, Q9UEF2, Q9UEG1, Q9UEK8, Q9UPK7 | Gene names | RPS27A, UBA80, UBCEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin. | |||||
|
UBIQ_MOUSE
|
||||||
| NC score | 0.313814 (rank : 12) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62991, P02248, P02249, P02250, Q29120, Q62317, Q64223, Q8VCH1, Q91887, Q91888, Q9CXY4, Q9CZM0, Q9D1R5, Q9D8D9, Q9ET23, Q9ET24, Q9Z0H9 | Gene names | Rps27a, Uba80, Ubcep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin. | |||||
|
UBD_HUMAN
|
||||||
| NC score | 0.292203 (rank : 13) | θ value | 0.00869519 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15205, Q5SUK2, Q96EC7 | Gene names | UBD, FAT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin D (Ubiquitin-like protein FAT10) (Diubiquitin). | |||||
|
UBD_MOUSE
|
||||||
| NC score | 0.275241 (rank : 14) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63072, Q9WV10 | Gene names | Ubd, Fat10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin D (Ubiquitin-like protein FAT10) (Diubiquitin). | |||||
|
NEDD8_HUMAN
|
||||||
| NC score | 0.238571 (rank : 15) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15843, Q6LES6 | Gene names | NEDD8 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 precursor (Ubiquitin-like protein Nedd8) (Neddylin). | |||||
|
NEDD8_MOUSE
|
||||||
| NC score | 0.238300 (rank : 16) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29595 | Gene names | Nedd8, Nedd-8 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 precursor (Ubiquitin-like protein Nedd8) (Neddylin) (Neural precursor cell expressed developmentally down-regulated protein 8). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.234265 (rank : 17) | θ value | 0.00035302 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
UBL4A_HUMAN
|
||||||
| NC score | 0.230409 (rank : 18) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P11441 | Gene names | UBL4A, DXS254E, GDX, UBL4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 4A (Ubiquitin-like protein GDX). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.225453 (rank : 19) | θ value | 0.00102713 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
UCRP_MOUSE
|
||||||
| NC score | 0.213158 (rank : 20) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64339, Q8K0H3 | Gene names | Isg15, G1p2, Ucrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin cross-reactive protein precursor (Interferon-stimulated protein 15) (IP17). | |||||
|
PRKN2_MOUSE
|
||||||
| NC score | 0.195499 (rank : 21) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WVS6, Q9ES22, Q9ES23 | Gene names | Park2, Prkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN). | |||||
|
PRKN2_HUMAN
|
||||||
| NC score | 0.182590 (rank : 22) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
UBL4A_MOUSE
|
||||||
| NC score | 0.169598 (rank : 23) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P21126 | Gene names | Ubl4a, Gdx, Ubl4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 4A (Ubiquitin-like protein GDX). | |||||
|
TMUB2_MOUSE
|
||||||
| NC score | 0.149588 (rank : 24) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3V209, Q8K0X0, Q91YQ0, Q9D8C5 | Gene names | Tmub2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
RD23A_HUMAN
|
||||||
| NC score | 0.148158 (rank : 25) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P54725 | Gene names | RAD23A | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog A (hHR23A). | |||||
|
TMUB2_HUMAN
|
||||||
| NC score | 0.132310 (rank : 26) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q71RG4, Q8NDI2, Q9BPZ5, Q9HAG3 | Gene names | TMUB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
RD23A_MOUSE
|
||||||
| NC score | 0.121881 (rank : 27) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54726 | Gene names | Rad23a, Mhr23a | |||
|
Domain Architecture |

|
|||||
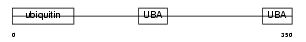
| Description | UV excision repair protein RAD23 homolog A (mHR23A). | |||||
|
RD23B_HUMAN
|
||||||
| NC score | 0.099437 (rank : 28) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P54727, Q8WUB0 | Gene names | RAD23B | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (hHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.095119 (rank : 29) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
RNF31_HUMAN
|
||||||
| NC score | 0.078996 (rank : 30) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |

|
|||||
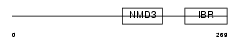
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
UBIM_HUMAN
|
||||||
| NC score | 0.075058 (rank : 31) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35544, Q9H5V4 | Gene names | FAU | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein FUBI. | |||||
|
SYRC_HUMAN
|
||||||
| NC score | 0.071136 (rank : 32) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54136, Q9BWA1 | Gene names | RARS | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA synthetase, cytoplasmic (EC 6.1.1.19) (Arginine--tRNA ligase) (ArgRS). | |||||
|
UBIM_MOUSE
|
||||||
| NC score | 0.070983 (rank : 33) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35545, Q9JJ24 | Gene names | Fau | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein FUBI (Monoclonal nonspecific suppressor factor beta) (MNSF-beta). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.069035 (rank : 34) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
RNF31_MOUSE
|
||||||
| NC score | 0.068078 (rank : 35) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
SYRC_MOUSE
|
||||||
| NC score | 0.062194 (rank : 36) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0I9, Q8VDW1 | Gene names | Rars | |||
|
Domain Architecture |
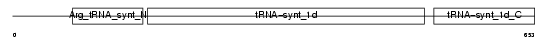
|
|||||
| Description | Arginyl-tRNA synthetase, cytoplasmic (EC 6.1.1.19) (Arginine--tRNA ligase) (ArgRS). | |||||
|
UCRP_HUMAN
|
||||||
| NC score | 0.059366 (rank : 37) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05161, Q7Z2G2, Q96GF0 | Gene names | ISG15, G1P2, UCRP | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced 17 kDa protein precursor [Contains: Ubiquitin cross-reactive protein (hUCRP) (Interferon-induced 15 kDa protein)]. | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.048311 (rank : 38) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.045753 (rank : 39) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.044977 (rank : 40) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.038604 (rank : 41) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SATB1_MOUSE
|
||||||
| NC score | 0.035475 (rank : 42) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60611 | Gene names | Satb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.035290 (rank : 43) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
SATB1_HUMAN
|
||||||
| NC score | 0.035119 (rank : 44) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01826 | Gene names | SATB1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.033946 (rank : 45) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
ADRM1_MOUSE
|
||||||
| NC score | 0.033646 (rank : 46) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
ADRM1_HUMAN
|
||||||
| NC score | 0.029742 (rank : 47) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.029125 (rank : 48) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.026777 (rank : 49) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.024909 (rank : 50) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.024693 (rank : 51) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
RINT1_MOUSE
|
||||||
| NC score | 0.022729 (rank : 52) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ36, Q3UXS1, Q6P1I2, Q8C193 | Gene names | Rint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD50-interacting protein 1 (RAD50 interactor 1) (Protein RINT-1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.021452 (rank : 53) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
DGCR8_MOUSE
|
||||||
| NC score | 0.021034 (rank : 54) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
YETS2_MOUSE
|
||||||
| NC score | 0.020875 (rank : 55) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.020847 (rank : 56) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
FOXP4_MOUSE
|
||||||
| NC score | 0.020611 (rank : 57) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
NUP62_MOUSE
|
||||||
| NC score | 0.020561 (rank : 58) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.019551 (rank : 59) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.019022 (rank : 60) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.018318 (rank : 61) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.017127 (rank : 62) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.016711 (rank : 63) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.016705 (rank : 64) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
THSD1_HUMAN
|
||||||
| NC score | 0.015822 (rank : 65) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.015789 (rank : 66) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.015639 (rank : 67) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
AMZ1_MOUSE
|
||||||
| NC score | 0.015162 (rank : 68) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVF9, Q811F9, Q8BMM5 | Gene names | Amz1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Archaemetzincin-1 (EC 3.-.-.-) (Archeobacterial metalloproteinase-like protein 1). | |||||
|
TDRD3_HUMAN
|
||||||
| NC score | 0.014315 (rank : 69) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.014180 (rank : 70) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
JHD2A_HUMAN
|
||||||
| NC score | 0.014155 (rank : 71) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.013962 (rank : 72) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.013143 (rank : 73) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.012979 (rank : 74) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
DPOE4_MOUSE
|
||||||
| NC score | 0.011292 (rank : 75) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQ36, Q3TJ13 | Gene names | Pole4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
RAD9A_MOUSE
|
||||||
| NC score | 0.010615 (rank : 76) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0F6, Q8QZZ3 | Gene names | Rad9a, Rad9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint control protein RAD9A (EC 3.1.11.2) (DNA repair exonuclease rad9 homolog A) (Rad9-like protein) (mRAD9). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.010471 (rank : 77) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.010442 (rank : 78) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.010148 (rank : 79) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
MBRL_HUMAN
|
||||||
| NC score | 0.009922 (rank : 80) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q4ZIN3, O60392, Q8NF79, Q96H30 | Gene names | C19orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membralin. | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.009508 (rank : 81) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.009446 (rank : 82) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HXD12_MOUSE
|
||||||
| NC score | 0.009091 (rank : 83) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23812 | Gene names | Hoxd12, Hox-4.7, Hoxd-12 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4.7) (Hox-5.6). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.008591 (rank : 84) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.008194 (rank : 85) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
K1849_HUMAN
|
||||||
| NC score | 0.008032 (rank : 86) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
DCC_HUMAN
|
||||||
| NC score | 0.006841 (rank : 87) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.006432 (rank : 88) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.006363 (rank : 89) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.006219 (rank : 90) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
DPEP3_MOUSE
|
||||||
| NC score | 0.006029 (rank : 91) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DA79 | Gene names | Dpep3, Mbd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidase 3 precursor (EC 3.4.13.19) (Membrane-bound dipeptidase 3) (MBD-3). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.004379 (rank : 92) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.003773 (rank : 93) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.003660 (rank : 94) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
IRAK1_MOUSE
|
||||||
| NC score | 0.003149 (rank : 95) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.002839 (rank : 96) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
KLF17_HUMAN
|
||||||
| NC score | 0.001976 (rank : 97) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
ISL2_HUMAN
|
||||||
| NC score | 0.001967 (rank : 98) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96A47 | Gene names | ISL2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
LRC8D_HUMAN
|
||||||
| NC score | 0.001852 (rank : 99) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L1W4, Q6UWB2, Q9NVW3 | Gene names | LRRC8D, LRRC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.001771 (rank : 100) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
KLF11_HUMAN
|
||||||
| NC score | 0.001408 (rank : 101) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14901 | Gene names | KLF11, FKLF, TIEG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 11 (Transforming growth factor-beta-inducible early growth response protein 2) (TGFB-inducible early growth response protein 2) (TIEG-2). | |||||
|
FATH_HUMAN
|
||||||
| NC score | -0.000475 (rank : 102) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||