Please be patient as the page loads
|
APLP1_MOUSE
|
||||||
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
APLP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.976296 (rank : 2) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP2_MOUSE
|
||||||
| θ value | 1.94503e-103 (rank : 3) | NC score | 0.902291 (rank : 3) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |
|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
APLP2_HUMAN
|
||||||
| θ value | 2.39424e-77 (rank : 4) | NC score | 0.812352 (rank : 4) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
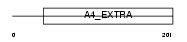
A4_HUMAN
|
||||||
| θ value | 6.76295e-64 (rank : 5) | NC score | 0.781049 (rank : 6) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_MOUSE
|
||||||
| θ value | 2.56992e-63 (rank : 6) | NC score | 0.781836 (rank : 5) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 7) | NC score | 0.011633 (rank : 166) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
AMOL2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 8) | NC score | 0.098997 (rank : 21) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 9) | NC score | 0.039871 (rank : 102) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 10) | NC score | 0.073890 (rank : 26) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 11) | NC score | 0.037384 (rank : 110) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 12) | NC score | 0.069515 (rank : 29) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.075936 (rank : 24) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
AMOL2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.086279 (rank : 23) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
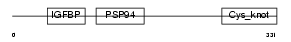
NOV_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.053371 (rank : 69) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.036190 (rank : 115) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.059018 (rank : 46) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.063227 (rank : 36) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
NTBP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.068525 (rank : 31) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.066184 (rank : 34) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.023125 (rank : 134) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.044042 (rank : 95) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.073940 (rank : 25) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.071701 (rank : 28) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.037715 (rank : 109) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
FES_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.008319 (rank : 176) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16879, Q62122 | Gene names | Fes, Fps | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.010458 (rank : 170) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.068511 (rank : 32) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.061270 (rank : 42) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.059209 (rank : 45) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.036117 (rank : 116) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.069172 (rank : 30) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.023127 (rank : 133) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.056423 (rank : 55) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
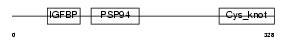
NOV_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.038168 (rank : 106) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.063149 (rank : 37) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CIP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.047896 (rank : 91) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.049078 (rank : 89) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
FA20A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.033648 (rank : 118) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CID3 | Gene names | Fam20a | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM20A precursor. | |||||
|
JHD3B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.013650 (rank : 159) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
M4K2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.012972 (rank : 160) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61161 | Gene names | Map4k2, Rab8ip | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GCK) (Rab8-interacting protein). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.058951 (rank : 47) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.026978 (rank : 126) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
GFAP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.036638 (rank : 113) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |
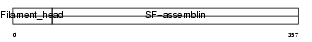
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.063367 (rank : 35) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
GP124_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.009230 (rank : 175) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.023408 (rank : 132) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.023558 (rank : 131) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
HRH1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.002802 (rank : 182) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70174, Q91V75, Q91XN0, Q91XN1, Q91XN2, Q91XN3 | Gene names | Hrh1, Bphs | |||
|
Domain Architecture |
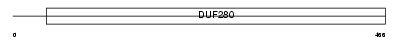
|
|||||
| Description | Histamine H1 receptor. | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.040677 (rank : 100) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
PLCB3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.028178 (rank : 122) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.056279 (rank : 56) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.049128 (rank : 88) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
LAMA5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.015947 (rank : 153) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.023035 (rank : 135) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.036962 (rank : 112) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.038093 (rank : 107) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.050618 (rank : 81) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
SNIP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.035511 (rank : 117) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.037373 (rank : 111) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
T3JAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.038528 (rank : 104) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.041751 (rank : 99) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.027404 (rank : 125) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.061744 (rank : 41) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
ARMC6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.028472 (rank : 121) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
AZI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.055408 (rank : 61) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.020787 (rank : 142) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DPOE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.023846 (rank : 130) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07864, Q13533, Q86VH9 | Gene names | POLE, POLE1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
K1718_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.014860 (rank : 155) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.036394 (rank : 114) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.057701 (rank : 52) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NUCB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.038227 (rank : 105) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.021516 (rank : 139) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.012410 (rank : 162) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
ADA11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.003812 (rank : 180) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
APC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.021345 (rank : 140) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.010021 (rank : 172) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.055545 (rank : 60) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.027437 (rank : 124) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.012332 (rank : 163) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.048232 (rank : 90) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DPOE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.022943 (rank : 136) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVF7, Q9QX50 | Gene names | Pole, Pole1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.063147 (rank : 38) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
HDAC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.042910 (rank : 98) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
JIP3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.037810 (rank : 108) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
LRFN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.003629 (rank : 181) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULH4, Q5SYP9 | Gene names | LRFN2, KIAA1246 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 2 precursor. | |||||
|
MPP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.010501 (rank : 168) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14168, Q9BQJ2 | Gene names | MPP2, DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Discs large homolog 2). | |||||
|
NHS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.022508 (rank : 138) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.017376 (rank : 148) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NPHP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.024823 (rank : 129) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.010390 (rank : 171) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.045771 (rank : 92) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.015919 (rank : 154) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
TAOK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.026566 (rank : 127) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
UBQL4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.009508 (rank : 174) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
ADA2B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | -0.000191 (rank : 183) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30545 | Gene names | Adra2b | |||
|
Domain Architecture |
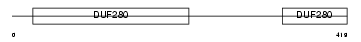
|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.006666 (rank : 178) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.027476 (rank : 123) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.044069 (rank : 94) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.030990 (rank : 120) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
EVC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.026231 (rank : 128) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P57679 | Gene names | EVC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein (DWF-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.018455 (rank : 145) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HOMEZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.011714 (rank : 165) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.013693 (rank : 158) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.038800 (rank : 103) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.017612 (rank : 147) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.006863 (rank : 177) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.045644 (rank : 93) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.021281 (rank : 141) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.019722 (rank : 144) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.018016 (rank : 146) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.043889 (rank : 96) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CCD27_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.043184 (rank : 97) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CCD39_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.040149 (rank : 101) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.055660 (rank : 58) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.006171 (rank : 179) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
GFAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.033100 (rank : 119) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |
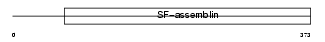
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
IL2RB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.010469 (rank : 169) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16297, Q3TZT2 | Gene names | Il2rb | |||
|
Domain Architecture |
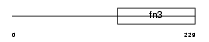
|
|||||
| Description | Interleukin-2 receptor beta chain precursor (IL-2 receptor) (P70-75) (High affinity IL-2 receptor subunit beta) (CD122 antigen). | |||||
|
K0157_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.013914 (rank : 157) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3TCJ1, Q3TB08, Q8K0R4 | Gene names | Kiaa0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
KI21B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.020344 (rank : 143) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.009939 (rank : 173) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MD2BP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.012778 (rank : 161) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCX1, Q99J05, Q9D0Y3 | Gene names | Mad2l1bp, Mad2lbp | |||
|
Domain Architecture |

|
|||||
| Description | MAD2L1-binding protein. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.016515 (rank : 149) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.022609 (rank : 137) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MPP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.011551 (rank : 167) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
SMG7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.016433 (rank : 150) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.016287 (rank : 151) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.014220 (rank : 156) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.011882 (rank : 164) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.016285 (rank : 152) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
AMBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.118758 (rank : 18) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |
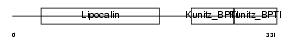
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
AMBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.120491 (rank : 16) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |
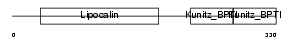
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
AMOL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.061928 (rank : 40) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IY63, Q63HK7, Q8NDN0, Q8WXD1, Q96CM5 | Gene names | AMOTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
AMOL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.055390 (rank : 62) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D4H4, Q571F1 | Gene names | Amotl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
AZI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.054695 (rank : 64) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
CA065_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.050364 (rank : 84) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.058477 (rank : 49) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.066398 (rank : 33) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.052202 (rank : 77) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.062889 (rank : 39) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.057307 (rank : 54) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CCD96_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.050001 (rank : 87) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CCD96_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.059482 (rank : 44) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.054659 (rank : 65) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CF060_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.073633 (rank : 27) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CING_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.053148 (rank : 70) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.055646 (rank : 59) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CO6A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.052551 (rank : 76) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
CP135_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.058779 (rank : 48) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP135_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.058262 (rank : 50) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DESP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.054803 (rank : 63) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
DREB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.051785 (rank : 79) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
EPPI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.154624 (rank : 10) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
EPPI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.142107 (rank : 12) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |
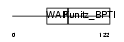
|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.052606 (rank : 75) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.054269 (rank : 67) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.054502 (rank : 66) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.050574 (rank : 82) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.058228 (rank : 51) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.050052 (rank : 86) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
INCE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.050473 (rank : 83) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INVO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.051788 (rank : 78) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.053116 (rank : 71) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.056236 (rank : 57) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.057525 (rank : 53) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.053002 (rank : 74) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.050240 (rank : 85) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.053508 (rank : 68) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.053023 (rank : 72) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SPIT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.105955 (rank : 20) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.108860 (rank : 19) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.160210 (rank : 9) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |
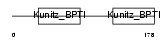
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
SPIT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.160702 (rank : 8) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |
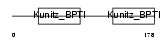
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
SPIT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.197923 (rank : 7) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
TFPI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.132145 (rank : 13) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |
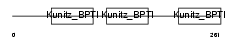
|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.128455 (rank : 14) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.118770 (rank : 17) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |
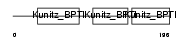
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
TFPI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.145862 (rank : 11) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |
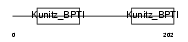
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.051694 (rank : 80) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TUFT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.053015 (rank : 73) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
WFDC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.089093 (rank : 22) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |
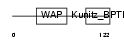
|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
WFDC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.122849 (rank : 15) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |
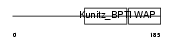
|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
ZWINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.060156 (rank : 43) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_HUMAN
|
||||||
| NC score | 0.976296 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP2_MOUSE
|
||||||
| NC score | 0.902291 (rank : 3) | θ value | 1.94503e-103 (rank : 3) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
APLP2_HUMAN
|
||||||
| NC score | 0.812352 (rank : 4) | θ value | 2.39424e-77 (rank : 4) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.781836 (rank : 5) | θ value | 2.56992e-63 (rank : 6) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.781049 (rank : 6) | θ value | 6.76295e-64 (rank : 5) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
SPIT3_HUMAN
|
||||||
| NC score | 0.197923 (rank : 7) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
SPIT2_MOUSE
|
||||||
| NC score | 0.160702 (rank : 8) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
SPIT2_HUMAN
|
||||||
| NC score | 0.160210 (rank : 9) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
EPPI_HUMAN
|
||||||
| NC score | 0.154624 (rank : 10) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
TFPI2_MOUSE
|
||||||
| NC score | 0.145862 (rank : 11) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
EPPI_MOUSE
|
||||||
| NC score | 0.142107 (rank : 12) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
TFPI1_HUMAN
|
||||||
| NC score | 0.132145 (rank : 13) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI1_MOUSE
|
||||||
| NC score | 0.128455 (rank : 14) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
WFDC8_HUMAN
|
||||||
| NC score | 0.122849 (rank : 15) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
AMBP_MOUSE
|
||||||
| NC score | 0.120491 (rank : 16) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
TFPI2_HUMAN
|
||||||
| NC score | 0.118770 (rank : 17) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
AMBP_HUMAN
|
||||||
| NC score | 0.118758 (rank : 18) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
SPIT1_MOUSE
|
||||||
| NC score | 0.108860 (rank : 19) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT1_HUMAN
|
||||||
| NC score | 0.105955 (rank : 20) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
AMOL2_HUMAN
|
||||||
| NC score | 0.098997 (rank : 21) | θ value | 0.00298849 (rank : 8) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
WFDC6_HUMAN
|
||||||
| NC score | 0.089093 (rank : 22) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
AMOL2_MOUSE
|
||||||
| NC score | 0.086279 (rank : 23) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.075936 (rank : 24) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.073940 (rank : 25) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.073890 (rank : 26) | θ value | 0.00665767 (rank : 10) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.073633 (rank : 27) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.071701 (rank : 28) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.069515 (rank : 29) | θ value | 0.00869519 (rank : 12) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.069172 (rank : 30) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
NTBP1_MOUSE
|
||||||
| NC score | 0.068525 (rank : 31) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.068511 (rank : 32) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.066398 (rank : 33) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.066184 (rank : 34) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.063367 (rank : 35) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.063227 (rank : 36) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.063149 (rank : 37) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.063147 (rank : 38) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.062889 (rank : 39) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
AMOL1_HUMAN
|
||||||
| NC score | 0.061928 (rank : 40) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IY63, Q63HK7, Q8NDN0, Q8WXD1, Q96CM5 | Gene names | AMOTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.061744 (rank : 41) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.061270 (rank : 42) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
ZWINT_MOUSE
|
||||||
| NC score | 0.060156 (rank : 43) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.059482 (rank : 44) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.059209 (rank : 45) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.059018 (rank : 46) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.058951 (rank : 47) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.058779 (rank : 48) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.058477 (rank : 49) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.058262 (rank : 50) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.058228 (rank : 51) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.057701 (rank : 52) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.057525 (rank : 53) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.057307 (rank : 54) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.056423 (rank : 55) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.056279 (rank : 56) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.056236 (rank : 57) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.055660 (rank : 58) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.055646 (rank : 59) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.055545 (rank : 60) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.055408 (rank : 61) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
AMOL1_MOUSE
|
||||||
| NC score | 0.055390 (rank : 62) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D4H4, Q571F1 | Gene names | Amotl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.054803 (rank : 63) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.054695 (rank : 64) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.054659 (rank : 65) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.054502 (rank : 66) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.054269 (rank : 67) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.053508 (rank : 68) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
NOV_MOUSE
|
||||||
| NC score | 0.053371 (rank : 69) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.053148 (rank : 70) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.053116 (rank : 71) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.053023 (rank : 72) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
TUFT1_MOUSE
|
||||||
| NC score | 0.053015 (rank : 73) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.053002 (rank : 74) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.052606 (rank : 75) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
CO6A3_HUMAN
|
||||||
| NC score | 0.052551 (rank : 76) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.052202 (rank : 77) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.051788 (rank : 78) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
DREB_HUMAN
|
||||||
| NC score | 0.051785 (rank : 79) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.051694 (rank : 80) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.050618 (rank : 81) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.050574 (rank : 82) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.050473 (rank : 83) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CA065_HUMAN
|
||||||
| NC score | 0.050364 (rank : 84) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.050240 (rank : 85) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.050052 (rank : 86) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
CCD96_HUMAN
|
||||||
| NC score | 0.050001 (rank : 87) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.049128 (rank : 88) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.049078 (rank : 89) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.048232 (rank : 90) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CIP4_HUMAN
|
||||||
| NC score | 0.047896 (rank : 91) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.045771 (rank : 92) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.045644 (rank : 93) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.044069 (rank : 94) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.044042 (rank : 95) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.043889 (rank : 96) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CCD27_HUMAN
|
||||||
| NC score | 0.043184 (rank : 97) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
HDAC4_HUMAN
|
||||||
| NC score | 0.042910 (rank : 98) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.041751 (rank : 99) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.040677 (rank : 100) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.040149 (rank : 101) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.039871 (rank : 102) | θ value | 0.00390308 (rank : 9) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.038800 (rank : 103) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
T3JAM_MOUSE
|
||||||
| NC score | 0.038528 (rank : 104) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
NUCB1_MOUSE
|
||||||
| NC score | 0.038227 (rank : 105) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
NOV_HUMAN
|
||||||
| NC score | 0.038168 (rank : 106) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.038093 (rank : 107) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
JIP3_MOUSE
|
||||||
| NC score | 0.037810 (rank : 108) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.037715 (rank : 109) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.037384 (rank : 110) | θ value | 0.00665767 (rank : 11) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.037373 (rank : 111) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.036962 (rank : 112) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
GFAP_MOUSE
|
||||||
| NC score | 0.036638 (rank : 113) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.036394 (rank : 114) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.036190 (rank : 115) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.036117 (rank : 116) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
SNIP_HUMAN
|
||||||
| NC score | 0.035511 (rank : 117) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
FA20A_MOUSE
|
||||||
| NC score | 0.033648 (rank : 118) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CID3 | Gene names | Fam20a | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM20A precursor. | |||||
|
GFAP_HUMAN
|
||||||
| NC score | 0.033100 (rank : 119) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.030990 (rank : 120) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ARMC6_MOUSE
|
||||||
| NC score | 0.028472 (rank : 121) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
PLCB3_MOUSE
|
||||||
| NC score | 0.028178 (rank : 122) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.027476 (rank : 123) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.027437 (rank : 124) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.027404 (rank : 125) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.026978 (rank : 126) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
TAOK1_MOUSE
|
||||||
| NC score | 0.026566 (rank : 127) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
EVC_HUMAN
|
||||||
| NC score | 0.026231 (rank : 128) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P57679 | Gene names | EVC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein (DWF-1). | |||||
|
NPHP1_HUMAN
|
||||||
| NC score | 0.024823 (rank : 129) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
DPOE1_HUMAN
|
||||||
| NC score | 0.023846 (rank : 130) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07864, Q13533, Q86VH9 | Gene names | POLE, POLE1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.023558 (rank : 131) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.023408 (rank : 132) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.023127 (rank : 133) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.023125 (rank : 134) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.023035 (rank : 135) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
DPOE1_MOUSE
|
||||||
| NC score | 0.022943 (rank : 136) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVF7, Q9QX50 | Gene names | Pole, Pole1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.022609 (rank : 137) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NHS_HUMAN
|
||||||
| NC score | 0.022508 (rank : 138) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.021516 (rank : 139) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.021345 (rank : 140) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.021281 (rank : 141) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.020787 (rank : 142) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
KI21B_HUMAN
|
||||||
| NC score | 0.020344 (rank : 143) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.019722 (rank : 144) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.018455 (rank : 145) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.018016 (rank : 146) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.017612 (rank : 147) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.017376 (rank : 148) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.016515 (rank : 149) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SMG7_HUMAN
|
||||||
| NC score | 0.016433 (rank : 150) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_MOUSE
|
||||||
| NC score | 0.016287 (rank : 151) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.016285 (rank : 152) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
LAMA5_HUMAN
|
||||||
| NC score | 0.015947 (rank : 153) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.015919 (rank : 154) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
K1718_HUMAN
|
||||||
| NC score | 0.014860 (rank : 155) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.014220 (rank : 156) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
K0157_MOUSE
|
||||||
| NC score | 0.013914 (rank : 157) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3TCJ1, Q3TB08, Q8K0R4 | Gene names | Kiaa0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.013693 (rank : 158) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
JHD3B_HUMAN
|
||||||
| NC score | 0.013650 (rank : 159) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
M4K2_MOUSE
|
||||||
| NC score | 0.012972 (rank : 160) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61161 | Gene names | Map4k2, Rab8ip | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GCK) (Rab8-interacting protein). | |||||
|
MD2BP_MOUSE
|
||||||
| NC score | 0.012778 (rank : 161) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCX1, Q99J05, Q9D0Y3 | Gene names | Mad2l1bp, Mad2lbp | |||
|
Domain Architecture |

|
|||||
| Description | MAD2L1-binding protein. | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.012410 (rank : 162) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.012332 (rank : 163) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.011882 (rank : 164) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
HOMEZ_HUMAN
|
||||||
| NC score | 0.011714 (rank : 165) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.011633 (rank : 166) | θ value | 0.00134147 (rank : 7) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
MPP2_MOUSE
|
||||||
| NC score | 0.011551 (rank : 167) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
MPP2_HUMAN
|
||||||
| NC score | 0.010501 (rank : 168) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14168, Q9BQJ2 | Gene names | MPP2, DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Discs large homolog 2). | |||||
|
IL2RB_MOUSE
|
||||||
| NC score | 0.010469 (rank : 169) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16297, Q3TZT2 | Gene names | Il2rb | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor beta chain precursor (IL-2 receptor) (P70-75) (High affinity IL-2 receptor subunit beta) (CD122 antigen). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.010458 (rank : 170) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.010390 (rank : 171) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | 0.010021 (rank : 172) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.009939 (rank : 173) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
UBQL4_MOUSE
|
||||||
| NC score | 0.009508 (rank : 174) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
GP124_MOUSE
|
||||||
| NC score | 0.009230 (rank : 175) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
FES_MOUSE
|
||||||
| NC score | 0.008319 (rank : 176) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16879, Q62122 | Gene names | Fes, Fps | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.006863 (rank : 177) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.006666 (rank : 178) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.006171 (rank : 179) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
ADA11_MOUSE
|
||||||
| NC score | 0.003812 (rank : 180) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
LRFN2_HUMAN
|
||||||
| NC score | 0.003629 (rank : 181) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULH4, Q5SYP9 | Gene names | LRFN2, KIAA1246 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 2 precursor. | |||||
|
HRH1_MOUSE
|
||||||
| NC score | 0.002802 (rank : 182) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70174, Q91V75, Q91XN0, Q91XN1, Q91XN2, Q91XN3 | Gene names | Hrh1, Bphs | |||
|
Domain Architecture |

|
|||||
| Description | Histamine H1 receptor. | |||||
|
ADA2B_MOUSE
|
||||||
| NC score | -0.000191 (rank : 183) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30545 | Gene names | Adra2b | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor). | |||||