Please be patient as the page loads
|
TFPI2_HUMAN
|
||||||
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |
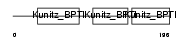
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TFPI2_HUMAN
|
||||||
| θ value | 6.01832e-121 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
TFPI2_MOUSE
|
||||||
| θ value | 2.17557e-62 (rank : 2) | NC score | 0.981915 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |
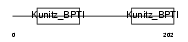
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
TFPI1_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 3) | NC score | 0.902584 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |
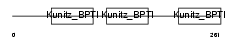
|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI1_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 4) | NC score | 0.888493 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
SPIT1_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 5) | NC score | 0.722076 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT1_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 6) | NC score | 0.697176 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT2_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 7) | NC score | 0.829780 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |
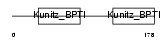
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
SPIT2_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 8) | NC score | 0.824458 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |
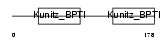
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
AMBP_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 9) | NC score | 0.674093 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |
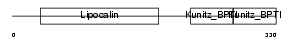
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
AMBP_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 10) | NC score | 0.685516 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |
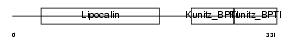
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
CO6A3_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 11) | NC score | 0.278543 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
EPPI_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 12) | NC score | 0.745481 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |
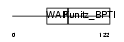
|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 13) | NC score | 0.146277 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
EPPI_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 14) | NC score | 0.723511 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
WFDC8_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 15) | NC score | 0.612926 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |
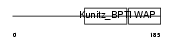
|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
A4_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 16) | NC score | 0.497613 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 17) | NC score | 0.498223 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
APLP2_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 18) | NC score | 0.492061 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
SPIT3_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 19) | NC score | 0.830543 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
WFDC6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.442519 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |
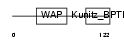
|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
EGFR_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.012335 (rank : 42) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00533, O00688, O00732, P06268, Q14225, Q92795, Q9BZS2, Q9GZX1, Q9H2C9, Q9H3C9, Q9UMD7, Q9UMD8, Q9UMG5 | Gene names | EGFR, ERBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1) (Receptor tyrosine-protein kinase ErbB-1). | |||||
|
ERBB4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.012881 (rank : 41) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15303 | Gene names | ERBB4, HER4 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-4 precursor (EC 2.7.10.1) (p180erbB4) (Tyrosine kinase-type cell surface receptor HER4). | |||||
|
EGFR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.010726 (rank : 43) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 927 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01279 | Gene names | Egfr | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1). | |||||
|
MEP1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.012893 (rank : 40) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28825 | Gene names | Mep1a | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (MEP-1). | |||||
|
ALK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.055145 (rank : 36) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03973, P07757 | Gene names | SLPI, WAP4, WFDC4 | |||
|
Domain Architecture |
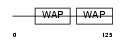
|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (HUSI-1) (Seminal proteinase inhibitor) (Secretory leukocyte protease inhibitor) (BLPI) (Mucus proteinase inhibitor) (MPI) (WAP four-disulfide core domain protein 4) (Protease inhibitor WAP4). | |||||
|
ALK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.061098 (rank : 34) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97430, O09081, O09082 | Gene names | Slpi | |||
|
Domain Architecture |
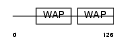
|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (Secretory leukocyte protease inhibitor). | |||||
|
APLP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.119011 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.118770 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.141380 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |
|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
CO8G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.084839 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
CO8G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.089643 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
ELAF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.050851 (rank : 39) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19957 | Gene names | PI3, WAP3, WFDC14 | |||
|
Domain Architecture |
|
|||||
| Description | Elafin precursor (Elastase-specific inhibitor) (ESI) (Skin-derived antileukoproteinase) (SKALP) (WAP four-disulfide core domain protein 14) (Protease inhibitor WAP3). | |||||
|
LRP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.109220 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
LRP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.109906 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
NGAL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.063167 (rank : 32) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P80188, P30150, Q92683 | Gene names | LCN2, HNL, NGAL | |||
|
Domain Architecture |
|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (25 kDa alpha-2-microglobulin-related subunit of MMP-9) (Lipocalin-2) (Oncogene 24p3). | |||||
|
PTGDS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.074756 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
PTGDS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.070611 (rank : 30) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||
|
VITRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.051086 (rank : 38) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
VITRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.051316 (rank : 37) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
WFD12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.061199 (rank : 33) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWY7, Q5H980, Q9BR31 | Gene names | WFDC12, C20orf122, WAP2 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Putative protease inhibitor WAP12) (Whey acidic protein 2). | |||||
|
WFD12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.068114 (rank : 31) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHY3 | Gene names | Wfdc12, Swam2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Whey acidic protein 2) (Single WAP motif protein 2) (Elafin-like protein II). | |||||
|
WFD15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.111230 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHY4, Q8BVC0 | Gene names | Wfdc15, Swam1 | |||
|
Domain Architecture |
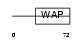
|
|||||
| Description | WAP four-disulfide core domain protein 15 precursor (Single WAP motif protein 1) (Elafin-like protein I). | |||||
|
WFDC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.056288 (rank : 35) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |
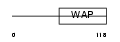
|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
TFPI2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.01832e-121 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
TFPI2_MOUSE
|
||||||
| NC score | 0.981915 (rank : 2) | θ value | 2.17557e-62 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
TFPI1_HUMAN
|
||||||
| NC score | 0.902584 (rank : 3) | θ value | 4.28545e-26 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI1_MOUSE
|
||||||
| NC score | 0.888493 (rank : 4) | θ value | 2.59989e-23 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
SPIT3_HUMAN
|
||||||
| NC score | 0.830543 (rank : 5) | θ value | 1.09739e-05 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
SPIT2_HUMAN
|
||||||
| NC score | 0.829780 (rank : 6) | θ value | 1.68911e-14 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
SPIT2_MOUSE
|
||||||
| NC score | 0.824458 (rank : 7) | θ value | 2.20605e-14 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
EPPI_MOUSE
|
||||||
| NC score | 0.745481 (rank : 8) | θ value | 7.34386e-10 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
EPPI_HUMAN
|
||||||
| NC score | 0.723511 (rank : 9) | θ value | 6.21693e-09 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
SPIT1_MOUSE
|
||||||
| NC score | 0.722076 (rank : 10) | θ value | 2.69047e-20 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT1_HUMAN
|
||||||
| NC score | 0.697176 (rank : 11) | θ value | 1.02238e-19 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
AMBP_HUMAN
|
||||||
| NC score | 0.685516 (rank : 12) | θ value | 2.0648e-12 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
AMBP_MOUSE
|
||||||
| NC score | 0.674093 (rank : 13) | θ value | 9.26847e-13 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
WFDC8_HUMAN
|
||||||
| NC score | 0.612926 (rank : 14) | θ value | 4.0297e-08 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.498223 (rank : 15) | θ value | 1.99992e-07 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.497613 (rank : 16) | θ value | 1.17247e-07 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
APLP2_HUMAN
|
||||||
| NC score | 0.492061 (rank : 17) | θ value | 3.41135e-07 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
WFDC6_HUMAN
|
||||||
| NC score | 0.442519 (rank : 18) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
CO6A3_HUMAN
|
||||||
| NC score | 0.278543 (rank : 19) | θ value | 8.67504e-11 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.146277 (rank : 20) | θ value | 4.76016e-09 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
APLP2_MOUSE
|
||||||
| NC score | 0.141380 (rank : 21) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
APLP1_HUMAN
|
||||||
| NC score | 0.119011 (rank : 22) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.118770 (rank : 23) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
WFD15_MOUSE
|
||||||
| NC score | 0.111230 (rank : 24) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHY4, Q8BVC0 | Gene names | Wfdc15, Swam1 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 15 precursor (Single WAP motif protein 1) (Elafin-like protein I). | |||||
|
LRP11_MOUSE
|
||||||
| NC score | 0.109906 (rank : 25) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
LRP11_HUMAN
|
||||||
| NC score | 0.109220 (rank : 26) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
CO8G_MOUSE
|
||||||
| NC score | 0.089643 (rank : 27) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
CO8G_HUMAN
|
||||||
| NC score | 0.084839 (rank : 28) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
PTGDS_HUMAN
|
||||||
| NC score | 0.074756 (rank : 29) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
PTGDS_MOUSE
|
||||||
| NC score | 0.070611 (rank : 30) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||
|
WFD12_MOUSE
|
||||||
| NC score | 0.068114 (rank : 31) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHY3 | Gene names | Wfdc12, Swam2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Whey acidic protein 2) (Single WAP motif protein 2) (Elafin-like protein II). | |||||
|
NGAL_HUMAN
|
||||||
| NC score | 0.063167 (rank : 32) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P80188, P30150, Q92683 | Gene names | LCN2, HNL, NGAL | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (25 kDa alpha-2-microglobulin-related subunit of MMP-9) (Lipocalin-2) (Oncogene 24p3). | |||||
|
WFD12_HUMAN
|
||||||
| NC score | 0.061199 (rank : 33) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWY7, Q5H980, Q9BR31 | Gene names | WFDC12, C20orf122, WAP2 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Putative protease inhibitor WAP12) (Whey acidic protein 2). | |||||
|
ALK1_MOUSE
|
||||||
| NC score | 0.061098 (rank : 34) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97430, O09081, O09082 | Gene names | Slpi | |||
|
Domain Architecture |

|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (Secretory leukocyte protease inhibitor). | |||||
|
WFDC5_HUMAN
|
||||||
| NC score | 0.056288 (rank : 35) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
ALK1_HUMAN
|
||||||
| NC score | 0.055145 (rank : 36) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03973, P07757 | Gene names | SLPI, WAP4, WFDC4 | |||
|
Domain Architecture |

|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (HUSI-1) (Seminal proteinase inhibitor) (Secretory leukocyte protease inhibitor) (BLPI) (Mucus proteinase inhibitor) (MPI) (WAP four-disulfide core domain protein 4) (Protease inhibitor WAP4). | |||||
|
VITRN_MOUSE
|
||||||
| NC score | 0.051316 (rank : 37) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
VITRN_HUMAN
|
||||||
| NC score | 0.051086 (rank : 38) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
ELAF_HUMAN
|
||||||
| NC score | 0.050851 (rank : 39) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19957 | Gene names | PI3, WAP3, WFDC14 | |||
|
Domain Architecture |

|
|||||
| Description | Elafin precursor (Elastase-specific inhibitor) (ESI) (Skin-derived antileukoproteinase) (SKALP) (WAP four-disulfide core domain protein 14) (Protease inhibitor WAP3). | |||||
|
MEP1A_MOUSE
|
||||||
| NC score | 0.012893 (rank : 40) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28825 | Gene names | Mep1a | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (MEP-1). | |||||
|
ERBB4_HUMAN
|
||||||
| NC score | 0.012881 (rank : 41) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15303 | Gene names | ERBB4, HER4 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-4 precursor (EC 2.7.10.1) (p180erbB4) (Tyrosine kinase-type cell surface receptor HER4). | |||||
|
EGFR_HUMAN
|
||||||
| NC score | 0.012335 (rank : 42) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00533, O00688, O00732, P06268, Q14225, Q92795, Q9BZS2, Q9GZX1, Q9H2C9, Q9H3C9, Q9UMD7, Q9UMD8, Q9UMG5 | Gene names | EGFR, ERBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1) (Receptor tyrosine-protein kinase ErbB-1). | |||||
|
EGFR_MOUSE
|
||||||
| NC score | 0.010726 (rank : 43) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 927 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01279 | Gene names | Egfr | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1). | |||||