Please be patient as the page loads
|
ALK1_HUMAN
|
||||||
| SwissProt Accessions | P03973, P07757 | Gene names | SLPI, WAP4, WFDC4 | |||
|
Domain Architecture |
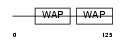
|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (HUSI-1) (Seminal proteinase inhibitor) (Secretory leukocyte protease inhibitor) (BLPI) (Mucus proteinase inhibitor) (MPI) (WAP four-disulfide core domain protein 4) (Protease inhibitor WAP4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ALK1_HUMAN
|
||||||
| θ value | 1.35638e-80 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P03973, P07757 | Gene names | SLPI, WAP4, WFDC4 | |||
|
Domain Architecture |

|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (HUSI-1) (Seminal proteinase inhibitor) (Secretory leukocyte protease inhibitor) (BLPI) (Mucus proteinase inhibitor) (MPI) (WAP four-disulfide core domain protein 4) (Protease inhibitor WAP4). | |||||
|
ALK1_MOUSE
|
||||||
| θ value | 1.08474e-45 (rank : 2) | NC score | 0.958277 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97430, O09081, O09082 | Gene names | Slpi | |||
|
Domain Architecture |
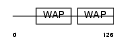
|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (Secretory leukocyte protease inhibitor). | |||||
|
WFDC5_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 3) | NC score | 0.757691 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |
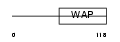
|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
WFDC3_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 4) | NC score | 0.570827 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
ELAF_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 5) | NC score | 0.742268 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19957 | Gene names | PI3, WAP3, WFDC14 | |||
|
Domain Architecture |
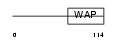
|
|||||
| Description | Elafin precursor (Elastase-specific inhibitor) (ESI) (Skin-derived antileukoproteinase) (SKALP) (WAP four-disulfide core domain protein 14) (Protease inhibitor WAP3). | |||||
|
WFDC2_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 6) | NC score | 0.644733 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14508, Q8WXV9, Q8WXW0, Q8WXW1, Q8WXW2, Q96KJ1 | Gene names | WFDC2, HE4, WAP5 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (Major epididymis- specific protein E4) (Epididymal secretory protein E4) (Putative protease inhibitor WAP5). | |||||
|
WFD12_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 7) | NC score | 0.725317 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWY7, Q5H980, Q9BR31 | Gene names | WFDC12, C20orf122, WAP2 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Putative protease inhibitor WAP12) (Whey acidic protein 2). | |||||
|
WFDC8_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 8) | NC score | 0.512662 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
WAP_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 9) | NC score | 0.593575 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P01173, P70230, Q61023 | Gene names | Wap | |||
|
Domain Architecture |
|
|||||
| Description | Whey acidic protein precursor (WAP). | |||||
|
WFD12_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 10) | NC score | 0.572379 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JHY3 | Gene names | Wfdc12, Swam2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Whey acidic protein 2) (Single WAP motif protein 2) (Elafin-like protein II). | |||||
|
EXPI_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 11) | NC score | 0.613751 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P62810, Q62477, Q91VQ6 | Gene names | Expi, Wdnm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular peptidase inhibitor precursor (Protein WDNM1). | |||||
|
EPPI_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 12) | NC score | 0.335641 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
EPPI_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 13) | NC score | 0.357675 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |
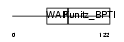
|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
WFDC2_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.550670 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DAU7 | Gene names | Wfdc2, He4 | |||
|
Domain Architecture |
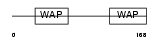
|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (WAP domain- containing protein HE4). | |||||
|
MCSP_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.211766 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
WFDC1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.289302 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HC57, Q8NC27, Q9HAU1 | Gene names | WFDC1, PS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 1 precursor (Prostate stromal protein ps20) (ps20 growth inhibitor). | |||||
|
WFDC1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.295520 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ESH5, Q8R110 | Gene names | Wfdc1, Ps20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 1 precursor (Prostate stromal protein ps20) (ps20 growth inhibitor). | |||||
|
WFDC6_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.327701 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |
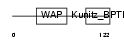
|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
WFD15_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.452294 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHY4, Q8BVC0 | Gene names | Wfdc15, Swam1 | |||
|
Domain Architecture |
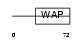
|
|||||
| Description | WAP four-disulfide core domain protein 15 precursor (Single WAP motif protein 1) (Elafin-like protein I). | |||||
|
KALM_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.284985 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23352 | Gene names | KAL1, ADMLX, KAL, KALIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Anosmin-1 precursor (Kallmann syndrome protein) (Adhesion molecule- like X-linked). | |||||
|
UROL1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.167741 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5DID3, Q5DID1, Q5DID2 | Gene names | Umodl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
UROL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.148923 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5DID0, Q5DIC9, Q6LA40, Q6LA41, Q8N216 | Gene names | UMODL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.056361 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.024197 (rank : 45) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.037404 (rank : 39) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.045147 (rank : 38) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
DLL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.026960 (rank : 43) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.032452 (rank : 40) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SORL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.021863 (rank : 47) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
TENA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.028772 (rank : 42) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.025278 (rank : 44) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.029434 (rank : 41) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
SORL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.022992 (rank : 46) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
KR171_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.052022 (rank : 35) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.059167 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.050797 (rank : 37) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.053981 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.056521 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.053188 (rank : 34) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
MCSP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.058191 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
SPIT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.051441 (rank : 36) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |
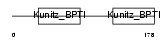
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
SPIT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.053524 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |
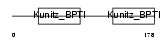
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
SPIT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.075123 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
TFPI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.055145 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |
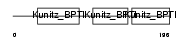
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
TFPI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.055493 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |
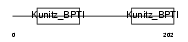
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
WF10A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.074831 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H1F0, Q5TGZ7 | Gene names | WFDC10A, C20orf146, WAP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 10A precursor (Putative protease inhibitor WAP10A). | |||||
|
WFD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.105151 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUB5, Q5TEU7, Q8WWK7 | Gene names | WFDC13, C20orf138, WAP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein WFDC13 precursor. | |||||
|
ALK1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.35638e-80 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P03973, P07757 | Gene names | SLPI, WAP4, WFDC4 | |||
|
Domain Architecture |

|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (HUSI-1) (Seminal proteinase inhibitor) (Secretory leukocyte protease inhibitor) (BLPI) (Mucus proteinase inhibitor) (MPI) (WAP four-disulfide core domain protein 4) (Protease inhibitor WAP4). | |||||
|
ALK1_MOUSE
|
||||||
| NC score | 0.958277 (rank : 2) | θ value | 1.08474e-45 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97430, O09081, O09082 | Gene names | Slpi | |||
|
Domain Architecture |

|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (Secretory leukocyte protease inhibitor). | |||||
|
WFDC5_HUMAN
|
||||||
| NC score | 0.757691 (rank : 3) | θ value | 9.26847e-13 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
ELAF_HUMAN
|
||||||
| NC score | 0.742268 (rank : 4) | θ value | 2.79066e-09 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19957 | Gene names | PI3, WAP3, WFDC14 | |||
|
Domain Architecture |

|
|||||
| Description | Elafin precursor (Elastase-specific inhibitor) (ESI) (Skin-derived antileukoproteinase) (SKALP) (WAP four-disulfide core domain protein 14) (Protease inhibitor WAP3). | |||||
|
WFD12_HUMAN
|
||||||
| NC score | 0.725317 (rank : 5) | θ value | 6.87365e-08 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWY7, Q5H980, Q9BR31 | Gene names | WFDC12, C20orf122, WAP2 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Putative protease inhibitor WAP12) (Whey acidic protein 2). | |||||
|
WFDC2_HUMAN
|
||||||
| NC score | 0.644733 (rank : 6) | θ value | 2.79066e-09 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14508, Q8WXV9, Q8WXW0, Q8WXW1, Q8WXW2, Q96KJ1 | Gene names | WFDC2, HE4, WAP5 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (Major epididymis- specific protein E4) (Epididymal secretory protein E4) (Putative protease inhibitor WAP5). | |||||
|
EXPI_MOUSE
|
||||||
| NC score | 0.613751 (rank : 7) | θ value | 0.000602161 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P62810, Q62477, Q91VQ6 | Gene names | Expi, Wdnm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular peptidase inhibitor precursor (Protein WDNM1). | |||||
|
WAP_MOUSE
|
||||||
| NC score | 0.593575 (rank : 8) | θ value | 1.87187e-05 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P01173, P70230, Q61023 | Gene names | Wap | |||
|
Domain Architecture |

|
|||||
| Description | Whey acidic protein precursor (WAP). | |||||
|
WFD12_MOUSE
|
||||||
| NC score | 0.572379 (rank : 9) | θ value | 5.44631e-05 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JHY3 | Gene names | Wfdc12, Swam2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Whey acidic protein 2) (Single WAP motif protein 2) (Elafin-like protein II). | |||||
|
WFDC3_HUMAN
|
||||||
| NC score | 0.570827 (rank : 10) | θ value | 3.89403e-11 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
WFDC2_MOUSE
|
||||||
| NC score | 0.550670 (rank : 11) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DAU7 | Gene names | Wfdc2, He4 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (WAP domain- containing protein HE4). | |||||
|
WFDC8_HUMAN
|
||||||
| NC score | 0.512662 (rank : 12) | θ value | 1.43324e-05 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
WFD15_MOUSE
|
||||||
| NC score | 0.452294 (rank : 13) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHY4, Q8BVC0 | Gene names | Wfdc15, Swam1 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 15 precursor (Single WAP motif protein 1) (Elafin-like protein I). | |||||
|
EPPI_MOUSE
|
||||||
| NC score | 0.357675 (rank : 14) | θ value | 0.00134147 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
EPPI_HUMAN
|
||||||
| NC score | 0.335641 (rank : 15) | θ value | 0.000786445 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
WFDC6_HUMAN
|
||||||
| NC score | 0.327701 (rank : 16) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
WFDC1_MOUSE
|
||||||
| NC score | 0.295520 (rank : 17) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ESH5, Q8R110 | Gene names | Wfdc1, Ps20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 1 precursor (Prostate stromal protein ps20) (ps20 growth inhibitor). | |||||
|
WFDC1_HUMAN
|
||||||
| NC score | 0.289302 (rank : 18) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HC57, Q8NC27, Q9HAU1 | Gene names | WFDC1, PS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 1 precursor (Prostate stromal protein ps20) (ps20 growth inhibitor). | |||||
|
KALM_HUMAN
|
||||||
| NC score | 0.284985 (rank : 19) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23352 | Gene names | KAL1, ADMLX, KAL, KALIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Anosmin-1 precursor (Kallmann syndrome protein) (Adhesion molecule- like X-linked). | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.211766 (rank : 20) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
UROL1_MOUSE
|
||||||
| NC score | 0.167741 (rank : 21) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5DID3, Q5DID1, Q5DID2 | Gene names | Umodl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
UROL1_HUMAN
|
||||||
| NC score | 0.148923 (rank : 22) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5DID0, Q5DIC9, Q6LA40, Q6LA41, Q8N216 | Gene names | UMODL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
WFD13_HUMAN
|
||||||
| NC score | 0.105151 (rank : 23) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUB5, Q5TEU7, Q8WWK7 | Gene names | WFDC13, C20orf138, WAP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein WFDC13 precursor. | |||||
|
SPIT3_HUMAN
|
||||||
| NC score | 0.075123 (rank : 24) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
WF10A_HUMAN
|
||||||
| NC score | 0.074831 (rank : 25) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H1F0, Q5TGZ7 | Gene names | WFDC10A, C20orf146, WAP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 10A precursor (Putative protease inhibitor WAP10A). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.059167 (rank : 26) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
MCSP_HUMAN
|
||||||
| NC score | 0.058191 (rank : 27) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.056521 (rank : 28) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.056361 (rank : 29) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
TFPI2_MOUSE
|
||||||
| NC score | 0.055493 (rank : 30) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
TFPI2_HUMAN
|
||||||
| NC score | 0.055145 (rank : 31) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.053981 (rank : 32) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
SPIT2_MOUSE
|
||||||
| NC score | 0.053524 (rank : 33) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.053188 (rank : 34) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.052022 (rank : 35) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
SPIT2_HUMAN
|
||||||
| NC score | 0.051441 (rank : 36) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.050797 (rank : 37) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.045147 (rank : 38) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.037404 (rank : 39) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.032452 (rank : 40) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.029434 (rank : 41) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
TENA_MOUSE
|
||||||
| NC score | 0.028772 (rank : 42) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.026960 (rank : 43) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.025278 (rank : 44) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.024197 (rank : 45) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
SORL_MOUSE
|
||||||
| NC score | 0.022992 (rank : 46) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
SORL_HUMAN
|
||||||
| NC score | 0.021863 (rank : 47) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||