Please be patient as the page loads
|
APLP1_HUMAN
|
||||||
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
APLP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.976296 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP2_MOUSE
|
||||||
| θ value | 9.32813e-106 (rank : 3) | NC score | 0.905145 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |
|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
APLP2_HUMAN
|
||||||
| θ value | 1.15091e-71 (rank : 4) | NC score | 0.811934 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
A4_MOUSE
|
||||||
| θ value | 5.7252e-63 (rank : 5) | NC score | 0.777401 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_HUMAN
|
||||||
| θ value | 1.27544e-62 (rank : 6) | NC score | 0.776012 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 7) | NC score | 0.097906 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.080345 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.011538 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.043923 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.046834 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
AMOL2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.058995 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
NOV_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.050217 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.017273 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
CTGE5_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.026575 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.032163 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.043403 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.018306 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
FA13C_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.026362 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.044379 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.028627 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.033646 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
FA20A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.034220 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CID3 | Gene names | Fam20a | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM20A precursor. | |||||
|
GFAP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.024207 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.016639 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
NDE1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.025458 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
NOV_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.034979 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
TF2LX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.018071 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUE1 | Gene names | TGIF2LX, TGIFLX | |||
|
Domain Architecture |
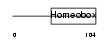
|
|||||
| Description | Homeobox protein TGIF2LX (TGFB-induced factor 2-like protein, X- linked) (TGF(beta)induced transcription factor 2-like protein) (TGIF- like on the X). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.028472 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.025362 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.026758 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRSK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.001497 (rank : 101) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.045607 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
CJ047_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.030827 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.014749 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.019827 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.021315 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.044943 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.025622 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CM007_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.016445 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.026011 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.019919 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.022825 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
IRAK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.003846 (rank : 99) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |
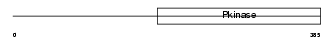
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.012367 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.018704 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TDRD6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.022894 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.022241 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ZN406_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.003489 (rank : 100) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P243, Q3MIM5, Q6PJ01, Q75PJ7, Q75PJ9, Q86X64 | Gene names | ZNF406, KIAA1485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406 (Protein ZFAT). | |||||
|
AMOL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.046314 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.024832 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.018017 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.009348 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
GGEE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.019294 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GU1, Q8WWL9 | Gene names | PAGE5, GAGEE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G antigen family E member 1 (Prostate-associated gene 5 protein) (PAGE-5). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.024083 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.053649 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.020257 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.025522 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
ARMC6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.026842 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.016038 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
GR65_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.014456 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
HMCS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.012396 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54868 | Gene names | HMGCS2 | |||
|
Domain Architecture |
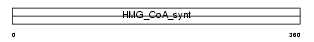
|
|||||
| Description | Hydroxymethylglutaryl-CoA synthase, mitochondrial precursor (EC 2.3.3.10) (HMG-CoA synthase) (3-hydroxy-3-methylglutaryl coenzyme A synthase). | |||||
|
MAGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.008443 (rank : 95) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |
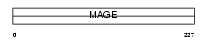
|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.029472 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.034656 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.039829 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
KI18A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.004179 (rank : 98) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NI77, Q86VS5, Q9H0F3 | Gene names | KIF18A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.010567 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.017535 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SNIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.027894 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.025569 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.022695 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.017564 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
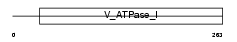
CRCM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.017260 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |
|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.006587 (rank : 97) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.023491 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
ERAL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.011017 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75616, O75617, Q8WUY4, Q96LE2, Q96TC0 | Gene names | ERAL1, HERA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (hERA) (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
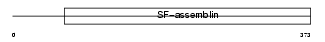
GFAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.019857 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
MPP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.006884 (rank : 96) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
MYCD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.008966 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
NAF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.028276 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.015671 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.025276 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.020764 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ZCHC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.012443 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
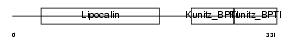
AMBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.118998 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
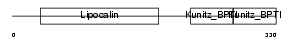
AMBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.120756 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
CO6A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.054507 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
EPPI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.154891 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
EPPI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.142363 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |
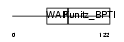
|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
SPIT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.106287 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.109135 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.160363 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |
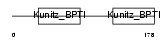
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
SPIT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.160900 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |
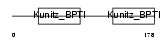
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
SPIT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.198096 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
TFPI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.132420 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |
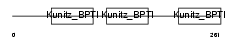
|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.128680 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.119011 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |
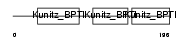
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
TFPI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.146076 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |
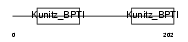
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
WFDC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.089189 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |
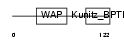
|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
WFDC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.123145 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |
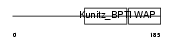
|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
APLP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.976296 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP2_MOUSE
|
||||||
| NC score | 0.905145 (rank : 3) | θ value | 9.32813e-106 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
APLP2_HUMAN
|
||||||
| NC score | 0.811934 (rank : 4) | θ value | 1.15091e-71 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.777401 (rank : 5) | θ value | 5.7252e-63 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.776012 (rank : 6) | θ value | 1.27544e-62 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
SPIT3_HUMAN
|
||||||
| NC score | 0.198096 (rank : 7) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
SPIT2_MOUSE
|
||||||
| NC score | 0.160900 (rank : 8) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
SPIT2_HUMAN
|
||||||
| NC score | 0.160363 (rank : 9) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
EPPI_HUMAN
|
||||||
| NC score | 0.154891 (rank : 10) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
TFPI2_MOUSE
|
||||||
| NC score | 0.146076 (rank : 11) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
EPPI_MOUSE
|
||||||
| NC score | 0.142363 (rank : 12) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
TFPI1_HUMAN
|
||||||
| NC score | 0.132420 (rank : 13) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI1_MOUSE
|
||||||
| NC score | 0.128680 (rank : 14) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
WFDC8_HUMAN
|
||||||
| NC score | 0.123145 (rank : 15) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
AMBP_MOUSE
|
||||||
| NC score | 0.120756 (rank : 16) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
TFPI2_HUMAN
|
||||||
| NC score | 0.119011 (rank : 17) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
AMBP_HUMAN
|
||||||
| NC score | 0.118998 (rank : 18) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
SPIT1_MOUSE
|
||||||
| NC score | 0.109135 (rank : 19) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT1_HUMAN
|
||||||
| NC score | 0.106287 (rank : 20) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.097906 (rank : 21) | θ value | 0.000270298 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
WFDC6_HUMAN
|
||||||
| NC score | 0.089189 (rank : 22) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.080345 (rank : 23) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
AMOL2_HUMAN
|
||||||
| NC score | 0.058995 (rank : 24) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
CO6A3_HUMAN
|
||||||
| NC score | 0.054507 (rank : 25) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.053649 (rank : 26) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
NOV_MOUSE
|
||||||
| NC score | 0.050217 (rank : 27) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.046834 (rank : 28) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
AMOL2_MOUSE
|
||||||
| NC score | 0.046314 (rank : 29) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.045607 (rank : 30) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.044943 (rank : 31) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.044379 (rank : 32) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.043923 (rank : 33) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.043403 (rank : 34) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.039829 (rank : 35) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
NOV_HUMAN
|
||||||
| NC score | 0.034979 (rank : 36) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.034656 (rank : 37) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
FA20A_MOUSE
|
||||||
| NC score | 0.034220 (rank : 38) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CID3 | Gene names | Fam20a | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM20A precursor. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.033646 (rank : 39) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.032163 (rank : 40) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
CJ047_MOUSE
|
||||||
| NC score | 0.030827 (rank : 41) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.029472 (rank : 42) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.028627 (rank : 43) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.028472 (rank : 44) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.028276 (rank : 45) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
SNIP_HUMAN
|
||||||
| NC score | 0.027894 (rank : 46) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
ARMC6_MOUSE
|
||||||
| NC score | 0.026842 (rank : 47) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.026758 (rank : 48) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
CTGE5_MOUSE
|
||||||
| NC score | 0.026575 (rank : 49) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
FA13C_MOUSE
|
||||||
| NC score | 0.026362 (rank : 50) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.026011 (rank : 51) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.025622 (rank : 52) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.025569 (rank : 53) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.025522 (rank : 54) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
NDE1_MOUSE
|
||||||
| NC score | 0.025458 (rank : 55) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.025362 (rank : 56) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.025276 (rank : 57) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.024832 (rank : 58) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
GFAP_MOUSE
|
||||||
| NC score | 0.024207 (rank : 59) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.024083 (rank : 60) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.023491 (rank : 61) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
TDRD6_MOUSE
|
||||||
| NC score | 0.022894 (rank : 62) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.022825 (rank : 63) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.022695 (rank : 64) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.022241 (rank : 65) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.021315 (rank : 66) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.020764 (rank : 67) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.020257 (rank : 68) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.019919 (rank : 69) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GFAP_HUMAN
|
||||||
| NC score | 0.019857 (rank : 70) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.019827 (rank : 71) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
GGEE1_HUMAN
|
||||||
| NC score | 0.019294 (rank : 72) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GU1, Q8WWL9 | Gene names | PAGE5, GAGEE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G antigen family E member 1 (Prostate-associated gene 5 protein) (PAGE-5). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.018704 (rank : 73) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.018306 (rank : 74) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
TF2LX_HUMAN
|
||||||
| NC score | 0.018071 (rank : 75) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUE1 | Gene names | TGIF2LX, TGIFLX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein TGIF2LX (TGFB-induced factor 2-like protein, X- linked) (TGF(beta)induced transcription factor 2-like protein) (TGIF- like on the X). | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.018017 (rank : 76) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.017564 (rank : 77) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.017535 (rank : 78) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.017273 (rank : 79) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
CRCM_HUMAN
|
||||||
| NC score | 0.017260 (rank : 80) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |

|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.016639 (rank : 81) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.016445 (rank : 82) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.016038 (rank : 83) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.015671 (rank : 84) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.014749 (rank : 85) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.014456 (rank : 86) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
ZCHC8_HUMAN
|
||||||
| NC score | 0.012443 (rank : 87) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
HMCS2_HUMAN
|
||||||
| NC score | 0.012396 (rank : 88) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54868 | Gene names | HMGCS2 | |||
|
Domain Architecture |
|
|||||
| Description | Hydroxymethylglutaryl-CoA synthase, mitochondrial precursor (EC 2.3.3.10) (HMG-CoA synthase) (3-hydroxy-3-methylglutaryl coenzyme A synthase). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.012367 (rank : 89) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.011538 (rank : 90) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
ERAL_HUMAN
|
||||||
| NC score | 0.011017 (rank : 91) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75616, O75617, Q8WUY4, Q96LE2, Q96TC0 | Gene names | ERAL1, HERA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (hERA) (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.010567 (rank : 92) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.009348 (rank : 93) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
MYCD_HUMAN
|
||||||
| NC score | 0.008966 (rank : 94) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
MAGB1_HUMAN
|
||||||
| NC score | 0.008443 (rank : 95) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MPP2_MOUSE
|
||||||
| NC score | 0.006884 (rank : 96) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.006587 (rank : 97) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
KI18A_HUMAN
|
||||||
| NC score | 0.004179 (rank : 98) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NI77, Q86VS5, Q9H0F3 | Gene names | KIF18A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
IRAK1_MOUSE
|
||||||
| NC score | 0.003846 (rank : 99) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
ZN406_HUMAN
|
||||||
| NC score | 0.003489 (rank : 100) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P243, Q3MIM5, Q6PJ01, Q75PJ7, Q75PJ9, Q86X64 | Gene names | ZNF406, KIAA1485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406 (Protein ZFAT). | |||||
|
BRSK1_HUMAN
|
||||||
| NC score | 0.001497 (rank : 101) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||