Please be patient as the page loads
|
ZCHC8_HUMAN
|
||||||
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZCHC8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.945976 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 3) | NC score | 0.081118 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 4) | NC score | 0.092945 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.078917 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RANB3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.069015 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.052645 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SF3B2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.133140 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.051063 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.032931 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.026101 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.061743 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CF010_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.067049 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.050996 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.048879 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.037488 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RPGR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.036625 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
ZCHC6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.061942 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.017686 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.039861 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
OGFR_HUMAN
|
||||||
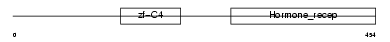
| θ value | 0.813845 (rank : 21) | NC score | 0.064599 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.044952 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.039397 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.032895 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
RBM4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.056206 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.056148 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.048244 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.039625 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.049837 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.036345 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.034091 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.056995 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.025931 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.044685 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RBM4B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.052641 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.052596 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
ATS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.015143 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHI8, Q9NSJ8, Q9P2K0, Q9UH83, Q9UP80 | Gene names | ADAMTS1, KIAA1346, METH1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1) (METH-1). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.029557 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.042506 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.034770 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.025256 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
SHOX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.009635 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15266, O00412, O00413, O15267 | Gene names | SHOX, PHOG | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein (Short stature homeobox-containing protein) (Pseudoautosomal homeobox-containing osteogenic protein). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.024462 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
CJ084_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.037327 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8W3, Q9H5V5 | Gene names | C10orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf84. | |||||
|
NDEL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.044797 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
NDEL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.044573 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
RXRA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.009263 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28700 | Gene names | Rxra, Nr2b1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.044653 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.032054 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
FA29A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.033673 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.041430 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
LN28B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.060525 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZN17 | Gene names | LIN28B, CSDD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.059582 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.037672 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.038889 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.026304 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.024602 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.041311 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.030685 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD91_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.035725 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.028518 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.028859 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.026772 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
SYRC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.019281 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0I9, Q8VDW1 | Gene names | Rars | |||
|
Domain Architecture |
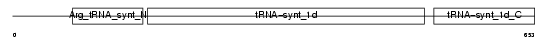
|
|||||
| Description | Arginyl-tRNA synthetase, cytoplasmic (EC 6.1.1.19) (Arginine--tRNA ligase) (ArgRS). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.035433 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.045955 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ZCHC9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.032162 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
ZFYV1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.015309 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
LN28A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.057178 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H9Z2 | Gene names | LIN28, CSDD1, LIN28A, ZCCHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Zinc finger CCHC domain-containing protein 1). | |||||
|
LN28A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.057493 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3Y3, Q6NV62 | Gene names | Lin28, Lin28a, Tex17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Testis-expressed protein 17). | |||||
|
PRGC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.022325 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
RXRA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.007818 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.034686 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.009815 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.028883 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.041630 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.032480 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ZN692_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.000280 (rank : 93) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
AL2S8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.014951 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHI4 | Gene names | Als2cr8, Carf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 8 protein homolog (Calcium-response factor) (CaRF). | |||||
|
APLP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.012443 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.030777 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.024428 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.006797 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.010977 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
MCES_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.016269 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0L8, Q3V3U9, Q6ZQC6, Q9D5F1 | Gene names | Rnmt, Kiaa0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase). | |||||
|
NDC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.014632 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCB1 | Gene names | Tmem48, Ndc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NDC1 (Transmembrane protein 48). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.025319 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.010741 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.025000 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
SCAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.009561 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
ZCH13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.025702 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
ZDHC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.009545 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WTX9, O15461 | Gene names | ZDHHC1, C16orf1, ZNF377 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1) (Zinc finger protein 377) (DHHC- domain-containing cysteine-rich protein 1). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.053646 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
ZCHC8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.945976 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
SF3B2_HUMAN
|
||||||
| NC score | 0.133140 (rank : 3) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.092945 (rank : 4) | θ value | 0.0193708 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.081118 (rank : 5) | θ value | 0.000461057 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.078917 (rank : 6) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RANB3_MOUSE
|
||||||
| NC score | 0.069015 (rank : 7) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.067049 (rank : 8) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
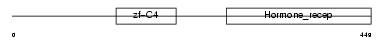
OGFR_HUMAN
|
||||||
| NC score | 0.064599 (rank : 9) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ZCHC6_HUMAN
|
||||||
| NC score | 0.061942 (rank : 10) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.061743 (rank : 11) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LN28B_HUMAN
|
||||||
| NC score | 0.060525 (rank : 12) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZN17 | Gene names | LIN28B, CSDD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28B_MOUSE
|
||||||
| NC score | 0.059582 (rank : 13) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28A_MOUSE
|
||||||
| NC score | 0.057493 (rank : 14) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3Y3, Q6NV62 | Gene names | Lin28, Lin28a, Tex17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Testis-expressed protein 17). | |||||
|
LN28A_HUMAN
|
||||||
| NC score | 0.057178 (rank : 15) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H9Z2 | Gene names | LIN28, CSDD1, LIN28A, ZCCHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Zinc finger CCHC domain-containing protein 1). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.056995 (rank : 16) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.056206 (rank : 17) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| NC score | 0.056148 (rank : 18) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.053646 (rank : 19) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.052645 (rank : 20) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RBM4B_HUMAN
|
||||||
| NC score | 0.052641 (rank : 21) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| NC score | 0.052596 (rank : 22) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.051063 (rank : 23) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.050996 (rank : 24) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.049837 (rank : 25) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.048879 (rank : 26) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.048244 (rank : 27) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.045955 (rank : 28) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.044952 (rank : 29) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
NDEL1_HUMAN
|
||||||
| NC score | 0.044797 (rank : 30) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.044685 (rank : 31) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.044653 (rank : 32) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
NDEL1_MOUSE
|
||||||
| NC score | 0.044573 (rank : 33) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.042506 (rank : 34) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.041630 (rank : 35) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.041430 (rank : 36) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.041311 (rank : 37) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.039861 (rank : 38) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.039625 (rank : 39) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.039397 (rank : 40) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.038889 (rank : 41) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.037672 (rank : 42) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.037488 (rank : 43) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CJ084_HUMAN
|
||||||
| NC score | 0.037327 (rank : 44) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8W3, Q9H5V5 | Gene names | C10orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf84. | |||||
|
RPGR_HUMAN
|
||||||
| NC score | 0.036625 (rank : 45) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.036345 (rank : 46) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CCD91_MOUSE
|
||||||
| NC score | 0.035725 (rank : 47) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.035433 (rank : 48) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.034770 (rank : 49) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.034686 (rank : 50) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.034091 (rank : 51) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
FA29A_HUMAN
|
||||||
| NC score | 0.033673 (rank : 52) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.032931 (rank : 53) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.032895 (rank : 54) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.032480 (rank : 55) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ZCHC9_HUMAN
|
||||||
| NC score | 0.032162 (rank : 56) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.032054 (rank : 57) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.030777 (rank : 58) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.030685 (rank : 59) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.029557 (rank : 60) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.028883 (rank : 61) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.028859 (rank : 62) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.028518 (rank : 63) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.026772 (rank : 64) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.026304 (rank : 65) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.026101 (rank : 66) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.025931 (rank : 67) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ZCH13_HUMAN
|
||||||
| NC score | 0.025702 (rank : 68) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.025319 (rank : 69) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.025256 (rank : 70) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.025000 (rank : 71) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.024602 (rank : 72) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.024462 (rank : 73) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.024428 (rank : 74) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
PRGC1_MOUSE
|
||||||
| NC score | 0.022325 (rank : 75) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
SYRC_MOUSE
|
||||||
| NC score | 0.019281 (rank : 76) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0I9, Q8VDW1 | Gene names | Rars | |||
|
Domain Architecture |
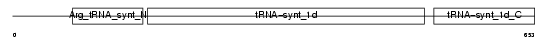
|
|||||
| Description | Arginyl-tRNA synthetase, cytoplasmic (EC 6.1.1.19) (Arginine--tRNA ligase) (ArgRS). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.017686 (rank : 77) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
MCES_MOUSE
|
||||||
| NC score | 0.016269 (rank : 78) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0L8, Q3V3U9, Q6ZQC6, Q9D5F1 | Gene names | Rnmt, Kiaa0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase). | |||||
|
ZFYV1_HUMAN
|
||||||
| NC score | 0.015309 (rank : 79) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
ATS1_HUMAN
|
||||||
| NC score | 0.015143 (rank : 80) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHI8, Q9NSJ8, Q9P2K0, Q9UH83, Q9UP80 | Gene names | ADAMTS1, KIAA1346, METH1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1) (METH-1). | |||||
|
AL2S8_MOUSE
|
||||||
| NC score | 0.014951 (rank : 81) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHI4 | Gene names | Als2cr8, Carf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 8 protein homolog (Calcium-response factor) (CaRF). | |||||
|
NDC1_MOUSE
|
||||||
| NC score | 0.014632 (rank : 82) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCB1 | Gene names | Tmem48, Ndc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NDC1 (Transmembrane protein 48). | |||||
|
APLP1_HUMAN
|
||||||
| NC score | 0.012443 (rank : 83) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.010977 (rank : 84) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.010741 (rank : 85) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.009815 (rank : 86) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SHOX_HUMAN
|
||||||
| NC score | 0.009635 (rank : 87) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15266, O00412, O00413, O15267 | Gene names | SHOX, PHOG | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein (Short stature homeobox-containing protein) (Pseudoautosomal homeobox-containing osteogenic protein). | |||||
|
SCAP_HUMAN
|
||||||
| NC score | 0.009561 (rank : 88) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
ZDHC1_HUMAN
|
||||||
| NC score | 0.009545 (rank : 89) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WTX9, O15461 | Gene names | ZDHHC1, C16orf1, ZNF377 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1) (Zinc finger protein 377) (DHHC- domain-containing cysteine-rich protein 1). | |||||
|
RXRA_MOUSE
|
||||||
| NC score | 0.009263 (rank : 90) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28700 | Gene names | Rxra, Nr2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
RXRA_HUMAN
|
||||||
| NC score | 0.007818 (rank : 91) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.006797 (rank : 92) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
ZN692_MOUSE
|
||||||
| NC score | 0.000280 (rank : 93) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||