Please be patient as the page loads
|
ZCHC8_MOUSE
|
||||||
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZCHC8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.945976 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 3) | NC score | 0.096797 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 4) | NC score | 0.099924 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 5) | NC score | 0.062515 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ZN206_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.007920 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.072025 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.050506 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SF3B2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.146651 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.043198 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
APBB2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.057487 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.077666 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TSH2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.035932 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.053784 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.047247 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.048854 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.049843 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.041006 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.054922 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.079914 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.067732 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.074789 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.030619 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.021605 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.051566 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
RBM4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.064318 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.064452 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
ATS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.017154 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHI8, Q9NSJ8, Q9P2K0, Q9UH83, Q9UP80 | Gene names | ADAMTS1, KIAA1346, METH1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1) (METH-1). | |||||
|
BRSK2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.004461 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.070043 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.053020 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.090582 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
DAB2P_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.044134 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.047278 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.046017 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
RBM4B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.060967 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.060910 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.023309 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
LN28A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.079602 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9Z2 | Gene names | LIN28, CSDD1, LIN28A, ZCCHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Zinc finger CCHC domain-containing protein 1). | |||||
|
LN28A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.079843 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3Y3, Q6NV62 | Gene names | Lin28, Lin28a, Tex17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Testis-expressed protein 17). | |||||
|
LN28B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.082846 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZN17 | Gene names | LIN28B, CSDD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.081472 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.050020 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.024239 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
BORG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.034835 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14613, Q9UNS0 | Gene names | CDC42EP2, BORG1, CEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 2 (Binder of Rho GTPases 1). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.042246 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.047424 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.043632 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.030218 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.077687 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.045278 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.045269 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.076900 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.038070 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.055789 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.037702 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
DAB2P_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.041812 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.050640 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.059947 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.044284 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.062798 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.065351 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.053811 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
DEAF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.033711 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.040365 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.030011 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.055480 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.042652 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
SON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.044748 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.036750 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.082333 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ZCHC6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.038196 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.052744 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.039781 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.027071 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
DNMBP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.013946 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.027618 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
FBX28_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.040348 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NVF7, O75070 | Gene names | FBXO28, KIAA0483 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 28. | |||||
|
GAB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.043231 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.028861 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
MPP10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.056481 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.033125 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
ZFP13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.002743 (rank : 163) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.035880 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
DMRTA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.022757 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VZB9, Q8N8Y9, Q9H4B9 | Gene names | DMRTA1, DMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor A1. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.038752 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.014544 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MBTP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.019315 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14703, Q9UF67 | Gene names | MBTPS1, KIAA0091, S1P, SKI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin- isozyme 1) (SKI-1). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.031697 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.031465 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
ZN710_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.003168 (rank : 162) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.029431 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
ECM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.025087 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
ECT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.010443 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.053977 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K0841_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.022356 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D786, Q5HZI7, Q6ZQ35, Q8CIK2 | Gene names | Kiaa0841 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0841. | |||||
|
K1543_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.041606 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.024943 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
MCM4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.010374 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.044248 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.035566 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NDEL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.039393 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
NDEL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.039531 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
OST3B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.008123 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y662 | Gene names | HS3ST3B1, 3OST3B1, HS3ST3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (h3-OST-3B). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.027056 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
SCNNA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.010878 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P37088, O43271, Q9UM64 | Gene names | SCNN1A, SCNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SLY_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.012897 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K352, Q3TBX0, Q9DBV2 | Gene names | Sly | |||
|
Domain Architecture |
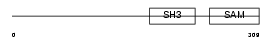
|
|||||
| Description | SH3 protein expressed in lymphocytes. | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.035204 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.040189 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.002247 (rank : 164) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.047063 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
BCL6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.003279 (rank : 161) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |
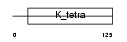
|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.024180 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
CF055_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.019154 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR26 | Gene names | ||||
|
Domain Architecture |
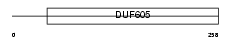
|
|||||
| Description | Protein C6orf55 homolog. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.033672 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.032936 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
DMRT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.026346 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5R5 | Gene names | DMRT2, DSXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Doublesex- and mab-3-related transcription factor 2 (Doublesex-like 2 protein) (DSXL-2). | |||||
|
DOK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.012824 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97465, Q9R213 | Gene names | Dok1, Dok | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.033930 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.032153 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
EXOC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.020047 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R3S6 | Gene names | Exoc1, Sec3, Sec3l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.010103 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
FAM5C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.013140 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76B58, O95726 | Gene names | FAM5C, DBCCR1L, DBCCR1L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5C precursor (DBCCR1-like protein 1). | |||||
|
FAM5C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.013147 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q499E0, Q8K0R9 | Gene names | Fam5c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5C precursor. | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.009815 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
IL16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.015052 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.045925 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.043757 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.018907 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.057014 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.055868 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RL6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.017215 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02878, Q2M3Q3, Q8WW97 | Gene names | RPL6 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L6 (TAX-responsive enhancer element-binding protein 107) (TAXREB107) (Neoplasm-related protein C140). | |||||
|
RON_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.000118 (rank : 165) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62190, Q62555 | Gene names | Mst1r, Ron, Stk | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (Stem cell-derived tyrosine kinase) (CDw136 antigen) [Contains: Macrophage-stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.033391 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.060106 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.051851 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.057153 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.053917 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.059436 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.056005 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.053661 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.069601 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.051513 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.051112 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EP300_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.053144 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.058413 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.068067 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.068772 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.052906 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.062449 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.052558 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.066897 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.068698 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.065384 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.052027 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.054706 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.056430 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.061898 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.053392 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.061790 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.063616 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.064454 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.050205 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.051526 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.050695 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
ZCHC8_HUMAN
|
||||||
| NC score | 0.945976 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
SF3B2_HUMAN
|
||||||
| NC score | 0.146651 (rank : 3) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.099924 (rank : 4) | θ value | 0.00298849 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.096797 (rank : 5) | θ value | 2.44474e-05 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.090582 (rank : 6) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
LN28B_HUMAN
|
||||||
| NC score | 0.082846 (rank : 7) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZN17 | Gene names | LIN28B, CSDD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.082333 (rank : 8) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
LN28B_MOUSE
|
||||||
| NC score | 0.081472 (rank : 9) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.079914 (rank : 10) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
LN28A_MOUSE
|
||||||
| NC score | 0.079843 (rank : 11) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3Y3, Q6NV62 | Gene names | Lin28, Lin28a, Tex17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Testis-expressed protein 17). | |||||
|
LN28A_HUMAN
|
||||||
| NC score | 0.079602 (rank : 12) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9Z2 | Gene names | LIN28, CSDD1, LIN28A, ZCCHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Zinc finger CCHC domain-containing protein 1). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.077687 (rank : 13) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.077666 (rank : 14) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.076900 (rank : 15) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.074789 (rank : 16) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.072025 (rank : 17) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.070043 (rank : 18) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.069601 (rank : 19) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.068772 (rank : 20) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.068698 (rank : 21) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.068067 (rank : 22) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.067732 (rank : 23) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.066897 (rank : 24) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.065384 (rank : 25) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.065351 (rank : 26) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.064454 (rank : 27) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
RBM4_MOUSE
|
||||||
| NC score | 0.064452 (rank : 28) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.064318 (rank : 29) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.063616 (rank : 30) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.062798 (rank : 31) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.062515 (rank : 32) | θ value | 0.00298849 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.062449 (rank : 33) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.061898 (rank : 34) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.061790 (rank : 35) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RBM4B_HUMAN
|
||||||
| NC score | 0.060967 (rank : 36) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| NC score | 0.060910 (rank : 37) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.060106 (rank : 38) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.059947 (rank : 39) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.059436 (rank : 40) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.058413 (rank : 41) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.057487 (rank : 42) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.057153 (rank : 43) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.057014 (rank : 44) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MPP10_HUMAN
|
||||||
| NC score | 0.056481 (rank : 45) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.056430 (rank : 46) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.056005 (rank : 47) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.055868 (rank : 48) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.055789 (rank : 49) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.055480 (rank : 50) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.054922 (rank : 51) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.054706 (rank : 52) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.053977 (rank : 53) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.053917 (rank : 54) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.053811 (rank : 55) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.053784 (rank : 56) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.053661 (rank : 57) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.053392 (rank : 58) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.053144 (rank : 59) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.053020 (rank : 60) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.052906 (rank : 61) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.052744 (rank : 62) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.052558 (rank : 63) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.052027 (rank : 64) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.051851 (rank : 65) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.051566 (rank : 66) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.051526 (rank : 67) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.051513 (rank : 68) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.051112 (rank : 69) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.050695 (rank : 70) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.050640 (rank : 71) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.050506 (rank : 72) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.050205 (rank : 73) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.050020 (rank : 74) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.049843 (rank : 75) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.048854 (rank : 76) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.047424 (rank : 77) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.047278 (rank : 78) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.047247 (rank : 79) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.047063 (rank : 80) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.046017 (rank : 81) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.045925 (rank : 82) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.045278 (rank : 83) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.045269 (rank : 84) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.044748 (rank : 85) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.044284 (rank : 86) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.044248 (rank : 87) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
DAB2P_MOUSE
|
||||||
| NC score | 0.044134 (rank : 88) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.043757 (rank : 89) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.043632 (rank : 90) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.043231 (rank : 91) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.043198 (rank : 92) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.042652 (rank : 93) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.042246 (rank : 94) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
DAB2P_HUMAN
|
||||||
| NC score | 0.041812 (rank : 95) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.041606 (rank : 96) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.041006 (rank : 97) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.040365 (rank : 98) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
FBX28_HUMAN
|
||||||
| NC score | 0.040348 (rank : 99) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NVF7, O75070 | Gene names | FBXO28, KIAA0483 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 28. | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.040189 (rank : 100) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.039781 (rank : 101) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
NDEL1_MOUSE
|
||||||
| NC score | 0.039531 (rank : 102) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
NDEL1_HUMAN
|
||||||
| NC score | 0.039393 (rank : 103) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.038752 (rank : 104) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
ZCHC6_HUMAN
|
||||||
| NC score | 0.038196 (rank : 105) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.038070 (rank : 106) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.037702 (rank : 107) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.036750 (rank : 108) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
TSH2_HUMAN
|
||||||
| NC score | 0.035932 (rank : 109) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.035880 (rank : 110) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.035566 (rank : 111) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.035204 (rank : 112) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
BORG1_HUMAN
|
||||||
| NC score | 0.034835 (rank : 113) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14613, Q9UNS0 | Gene names | CDC42EP2, BORG1, CEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 2 (Binder of Rho GTPases 1). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.033930 (rank : 114) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DEAF1_MOUSE
|
||||||
| NC score | 0.033711 (rank : 115) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.033672 (rank : 116) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.033391 (rank : 117) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.033125 (rank : 118) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.032936 (rank : 119) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.032153 (rank : 120) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.031697 (rank : 121) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.031465 (rank : 122) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.030619 (rank : 123) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.030218 (rank : 124) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.030011 (rank : 125) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.029431 (rank : 126) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.028861 (rank : 127) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.027618 (rank : 128) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.027071 (rank : 129) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.027056 (rank : 130) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
DMRT2_HUMAN
|
||||||
| NC score | 0.026346 (rank : 131) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5R5 | Gene names | DMRT2, DSXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Doublesex- and mab-3-related transcription factor 2 (Doublesex-like 2 protein) (DSXL-2). | |||||
|
ECM1_HUMAN
|
||||||
| NC score | 0.025087 (rank : 132) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.024943 (rank : 133) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.024239 (rank : 134) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.024180 (rank : 135) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.023309 (rank : 136) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
DMRTA_HUMAN
|
||||||
| NC score | 0.022757 (rank : 137) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VZB9, Q8N8Y9, Q9H4B9 | Gene names | DMRTA1, DMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor A1. | |||||
|
K0841_MOUSE
|
||||||
| NC score | 0.022356 (rank : 138) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D786, Q5HZI7, Q6ZQ35, Q8CIK2 | Gene names | Kiaa0841 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0841. | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.021605 (rank : 139) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
EXOC1_MOUSE
|
||||||
| NC score | 0.020047 (rank : 140) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R3S6 | Gene names | Exoc1, Sec3, Sec3l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
MBTP1_HUMAN
|
||||||
| NC score | 0.019315 (rank : 141) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14703, Q9UF67 | Gene names | MBTPS1, KIAA0091, S1P, SKI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin- isozyme 1) (SKI-1). | |||||
|
CF055_MOUSE
|
||||||
| NC score | 0.019154 (rank : 142) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR26 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C6orf55 homolog. | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.018907 (rank : 143) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RL6_HUMAN
|
||||||
| NC score | 0.017215 (rank : 144) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02878, Q2M3Q3, Q8WW97 | Gene names | RPL6 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L6 (TAX-responsive enhancer element-binding protein 107) (TAXREB107) (Neoplasm-related protein C140). | |||||
|
ATS1_HUMAN
|
||||||
| NC score | 0.017154 (rank : 145) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHI8, Q9NSJ8, Q9P2K0, Q9UH83, Q9UP80 | Gene names | ADAMTS1, KIAA1346, METH1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1) (METH-1). | |||||
|
IL16_HUMAN
|
||||||
| NC score | 0.015052 (rank : 146) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.014544 (rank : 147) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
DNMBP_HUMAN
|
||||||
| NC score | 0.013946 (rank : 148) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
FAM5C_MOUSE
|
||||||
| NC score | 0.013147 (rank : 149) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q499E0, Q8K0R9 | Gene names | Fam5c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5C precursor. | |||||
|
FAM5C_HUMAN
|
||||||
| NC score | 0.013140 (rank : 150) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76B58, O95726 | Gene names | FAM5C, DBCCR1L, DBCCR1L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5C precursor (DBCCR1-like protein 1). | |||||
|
SLY_MOUSE
|
||||||
| NC score | 0.012897 (rank : 151) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K352, Q3TBX0, Q9DBV2 | Gene names | Sly | |||
|
Domain Architecture |

|
|||||
| Description | SH3 protein expressed in lymphocytes. | |||||
|
DOK1_MOUSE
|
||||||
| NC score | 0.012824 (rank : 152) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97465, Q9R213 | Gene names | Dok1, Dok | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)). | |||||
|
SCNNA_HUMAN
|
||||||
| NC score | 0.010878 (rank : 153) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P37088, O43271, Q9UM64 | Gene names | SCNN1A, SCNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
ECT2_HUMAN
|
||||||
| NC score | 0.010443 (rank : 154) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
MCM4_HUMAN
|
||||||
| NC score | 0.010374 (rank : 155) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.010103 (rank : 156) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.009815 (rank : 157) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
OST3B_HUMAN
|
||||||
| NC score | 0.008123 (rank : 158) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y662 | Gene names | HS3ST3B1, 3OST3B1, HS3ST3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (h3-OST-3B). | |||||
|
ZN206_HUMAN
|
||||||
| NC score | 0.007920 (rank : 159) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||
|
BRSK2_HUMAN
|
||||||
| NC score | 0.004461 (rank : 160) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
BCL6_HUMAN
|
||||||
| NC score | 0.003279 (rank : 161) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||
|
ZN710_HUMAN
|
||||||
| NC score | 0.003168 (rank : 162) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
ZFP13_MOUSE
|
||||||
| NC score | 0.002743 (rank : 163) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.002247 (rank : 164) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
RON_MOUSE
|
||||||
| NC score | 0.000118 (rank : 165) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62190, Q62555 | Gene names | Mst1r, Ron, Stk | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (Stem cell-derived tyrosine kinase) (CDw136 antigen) [Contains: Macrophage-stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||