Please be patient as the page loads
|
BORG1_HUMAN
|
||||||
| SwissProt Accessions | O14613, Q9UNS0 | Gene names | CDC42EP2, BORG1, CEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 2 (Binder of Rho GTPases 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BORG1_HUMAN
|
||||||
| θ value | 5.09482e-120 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O14613, Q9UNS0 | Gene names | CDC42EP2, BORG1, CEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 2 (Binder of Rho GTPases 1). | |||||
|
BORG1_MOUSE
|
||||||
| θ value | 3.91911e-104 (rank : 2) | NC score | 0.983409 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8JZX9 | Gene names | Cdc42ep2, Borg1, Cep2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 2 (Binder of Rho GTPases 1). | |||||
|
BORG2_MOUSE
|
||||||
| θ value | 3.16345e-37 (rank : 3) | NC score | 0.892389 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQC5, Q3UD77, Q8BVR7 | Gene names | Cdc42ep3, Borg2, Cep3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2). | |||||
|
BORG2_HUMAN
|
||||||
| θ value | 5.0505e-35 (rank : 4) | NC score | 0.891020 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKI2, O95353, Q9UQJ0 | Gene names | CDC42EP3, BORG2, CEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2) (MSE55-related Cdc42-binding protein). | |||||
|
BORG5_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 5) | NC score | 0.673491 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |
|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
BORG4_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 6) | NC score | 0.682767 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM96, Q3TNF3, Q9QZT8 | Gene names | Cdc42ep4, Borg4, Cep4 | |||
|
Domain Architecture |
|
|||||
| Description | Cdc42 effector protein 4 (Binder of Rho GTPases 4). | |||||
|
BORG4_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 7) | NC score | 0.685475 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H3Q1, O95828, Q96FT3 | Gene names | CDC42EP4, BORG4, CEP4 | |||
|
Domain Architecture |
|
|||||
| Description | Cdc42 effector protein 4 (Binder of Rho GTPases 4). | |||||
|
BORG5_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 8) | NC score | 0.676451 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91W92, Q9D8M1 | Gene names | Cdc42ep1, Borg5, Cep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5). | |||||
|
BORG3_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 9) | NC score | 0.615372 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z0X0, Q3SYK1, Q9QZT9 | Gene names | Cdc42ep5, Borg3, Cep5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 5 (Binder of Rho GTPases 3). | |||||
|
BORG3_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 10) | NC score | 0.625600 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZY7, Q8TB51 | Gene names | CDC42EP5, BORG3, CEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 5 (Binder of Rho GTPases 3). | |||||
|
TGIF2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.051843 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZN2 | Gene names | TGIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein TGIF2 (TGFB-induced factor 2) (5'-TG-3'-interacting factor 2) (TGF(beta)-induced transcription factor 2). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.064941 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.036463 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
SP5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.010529 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.031806 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.046804 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MEF2D_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.025671 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63943, Q63944 | Gene names | Mef2d | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.013004 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
ZN341_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.006197 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYN7, Q5JXM8, Q96ST5 | Gene names | ZNF341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 341. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.037251 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.055623 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.054060 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.052662 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.006679 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKG_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.006973 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.034835 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.024734 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
ITB4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.011042 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.023324 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
DAG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.030089 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14118 | Gene names | DAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
SP5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.009080 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
TGIF2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.039886 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0Y1 | Gene names | Tgif2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein TGIF2 (TGFB-induced factor 2) (5'-TG-3'-interacting factor 2) (TGF(beta)-induced transcription factor 2). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.024907 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.009658 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.039260 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
FYB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.025049 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
HXA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.010143 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
RX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.009904 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
SON_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.049473 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.031531 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
IER5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.025287 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
NIBL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.030494 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96TA1, Q9BUS2, Q9NT35 | Gene names | C9orf88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein (Meg-3). | |||||
|
PAK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.004978 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13177, Q13154 | Gene names | PAK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 2 (EC 2.7.11.1) (p21-activated kinase 2) (PAK-2) (PAK65) (Gamma-PAK) (S6/H4 kinase). | |||||
|
PAK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.004985 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CIN4 | Gene names | Pak2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 2 (EC 2.7.11.1) (p21-activated kinase 2) (PAK-2). | |||||
|
RANB9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.025008 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.024642 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.049301 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.033026 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.020928 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.023782 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SOX8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.011339 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SRPK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.008129 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96SB4, Q12890, Q5R364, Q5R365, Q8IY12 | Gene names | SRPK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
ALEX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.021610 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.013795 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.011779 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
NIBL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.025946 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1F1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein. | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.005538 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.012994 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
SASH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.019240 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
SPN90_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.014428 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZQ3, Q9UGM8 | Gene names | NCKIPSD, AF3P21, SPIN90 | |||
|
Domain Architecture |
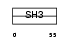
|
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (AF3p21) (Diaphanous protein-interacting protein) (Dia-interacting protein 1) (DIP-1). | |||||
|
ZN694_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.002079 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.007775 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.022796 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.005446 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.022248 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.013784 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.009712 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
BORG1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.09482e-120 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O14613, Q9UNS0 | Gene names | CDC42EP2, BORG1, CEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 2 (Binder of Rho GTPases 1). | |||||
|
BORG1_MOUSE
|
||||||
| NC score | 0.983409 (rank : 2) | θ value | 3.91911e-104 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8JZX9 | Gene names | Cdc42ep2, Borg1, Cep2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 2 (Binder of Rho GTPases 1). | |||||
|
BORG2_MOUSE
|
||||||
| NC score | 0.892389 (rank : 3) | θ value | 3.16345e-37 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQC5, Q3UD77, Q8BVR7 | Gene names | Cdc42ep3, Borg2, Cep3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2). | |||||
|
BORG2_HUMAN
|
||||||
| NC score | 0.891020 (rank : 4) | θ value | 5.0505e-35 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKI2, O95353, Q9UQJ0 | Gene names | CDC42EP3, BORG2, CEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2) (MSE55-related Cdc42-binding protein). | |||||
|
BORG4_HUMAN
|
||||||
| NC score | 0.685475 (rank : 5) | θ value | 4.16044e-13 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H3Q1, O95828, Q96FT3 | Gene names | CDC42EP4, BORG4, CEP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 4 (Binder of Rho GTPases 4). | |||||
|
BORG4_MOUSE
|
||||||
| NC score | 0.682767 (rank : 6) | θ value | 1.68911e-14 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM96, Q3TNF3, Q9QZT8 | Gene names | Cdc42ep4, Borg4, Cep4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 4 (Binder of Rho GTPases 4). | |||||
|
BORG5_MOUSE
|
||||||
| NC score | 0.676451 (rank : 7) | θ value | 1.02475e-11 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91W92, Q9D8M1 | Gene names | Cdc42ep1, Borg5, Cep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5). | |||||
|
BORG5_HUMAN
|
||||||
| NC score | 0.673491 (rank : 8) | θ value | 9.90251e-15 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
BORG3_HUMAN
|
||||||
| NC score | 0.625600 (rank : 9) | θ value | 1.69304e-06 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZY7, Q8TB51 | Gene names | CDC42EP5, BORG3, CEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 5 (Binder of Rho GTPases 3). | |||||
|
BORG3_MOUSE
|
||||||
| NC score | 0.615372 (rank : 10) | θ value | 9.92553e-07 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z0X0, Q3SYK1, Q9QZT9 | Gene names | Cdc42ep5, Borg3, Cep5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 5 (Binder of Rho GTPases 3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.064941 (rank : 11) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.055623 (rank : 12) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.054060 (rank : 13) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.052662 (rank : 14) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
TGIF2_HUMAN
|
||||||
| NC score | 0.051843 (rank : 15) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZN2 | Gene names | TGIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein TGIF2 (TGFB-induced factor 2) (5'-TG-3'-interacting factor 2) (TGF(beta)-induced transcription factor 2). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.049473 (rank : 16) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.049301 (rank : 17) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.046804 (rank : 18) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
TGIF2_MOUSE
|
||||||
| NC score | 0.039886 (rank : 19) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0Y1 | Gene names | Tgif2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein TGIF2 (TGFB-induced factor 2) (5'-TG-3'-interacting factor 2) (TGF(beta)-induced transcription factor 2). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.039260 (rank : 20) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.037251 (rank : 21) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.036463 (rank : 22) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.034835 (rank : 23) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.033026 (rank : 24) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.031806 (rank : 25) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.031531 (rank : 26) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
NIBL_HUMAN
|
||||||
| NC score | 0.030494 (rank : 27) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96TA1, Q9BUS2, Q9NT35 | Gene names | C9orf88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein (Meg-3). | |||||
|
DAG1_HUMAN
|
||||||
| NC score | 0.030089 (rank : 28) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14118 | Gene names | DAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
NIBL_MOUSE
|
||||||
| NC score | 0.025946 (rank : 29) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1F1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein. | |||||
|
MEF2D_MOUSE
|
||||||
| NC score | 0.025671 (rank : 30) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63943, Q63944 | Gene names | Mef2d | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
IER5_MOUSE
|
||||||
| NC score | 0.025287 (rank : 31) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.025049 (rank : 32) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
RANB9_HUMAN
|
||||||
| NC score | 0.025008 (rank : 33) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.024907 (rank : 34) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.024734 (rank : 35) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.024642 (rank : 36) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.023782 (rank : 37) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.023324 (rank : 38) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.022796 (rank : 39) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.022248 (rank : 40) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.021610 (rank : 41) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.020928 (rank : 42) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
SASH1_MOUSE
|
||||||
| NC score | 0.019240 (rank : 43) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
SPN90_HUMAN
|
||||||
| NC score | 0.014428 (rank : 44) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZQ3, Q9UGM8 | Gene names | NCKIPSD, AF3P21, SPIN90 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (AF3p21) (Diaphanous protein-interacting protein) (Dia-interacting protein 1) (DIP-1). | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.013795 (rank : 45) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.013784 (rank : 46) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.013004 (rank : 47) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.012994 (rank : 48) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.011779 (rank : 49) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
SOX8_HUMAN
|
||||||
| NC score | 0.011339 (rank : 50) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
ITB4_HUMAN
|
||||||
| NC score | 0.011042 (rank : 51) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.010529 (rank : 52) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
HXA3_MOUSE
|
||||||
| NC score | 0.010143 (rank : 53) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
RX_HUMAN
|
||||||
| NC score | 0.009904 (rank : 54) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.009712 (rank : 55) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | 0.009658 (rank : 56) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.009080 (rank : 57) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
SRPK1_HUMAN
|
||||||
| NC score | 0.008129 (rank : 58) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96SB4, Q12890, Q5R364, Q5R365, Q8IY12 | Gene names | SRPK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.007775 (rank : 59) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
MRCKG_MOUSE
|
||||||
| NC score | 0.006973 (rank : 60) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.006679 (rank : 61) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
ZN341_HUMAN
|
||||||
| NC score | 0.006197 (rank : 62) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYN7, Q5JXM8, Q96ST5 | Gene names | ZNF341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 341. | |||||
|
PKN3_MOUSE
|
||||||
| NC score | 0.005538 (rank : 63) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.005446 (rank : 64) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
PAK2_MOUSE
|
||||||
| NC score | 0.004985 (rank : 65) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CIN4 | Gene names | Pak2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 2 (EC 2.7.11.1) (p21-activated kinase 2) (PAK-2). | |||||
|
PAK2_HUMAN
|
||||||
| NC score | 0.004978 (rank : 66) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13177, Q13154 | Gene names | PAK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 2 (EC 2.7.11.1) (p21-activated kinase 2) (PAK-2) (PAK65) (Gamma-PAK) (S6/H4 kinase). | |||||
|
ZN694_HUMAN
|
||||||
| NC score | 0.002079 (rank : 67) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||