Please be patient as the page loads
|
MCM4_HUMAN
|
||||||
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MCM4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MCM4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995271 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P49717, O89056 | Gene names | Mcm4, Cdc21, Mcmd4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MCM2_HUMAN
|
||||||
| θ value | 5.30912e-93 (rank : 3) | NC score | 0.948537 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49736, Q14577, Q15023, Q8N2V1, Q969W7, Q96AE1, Q9BRM7 | Gene names | MCM2, BM28, CDCL1, KIAA0030 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM2 (Minichromosome maintenance protein 2 homolog) (Nuclear protein BM28). | |||||
|
MCM2_MOUSE
|
||||||
| θ value | 2.63489e-92 (rank : 4) | NC score | 0.947974 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97310, O08971, O89057, Q8C2R0 | Gene names | Mcm2, Bm28, Cdcl1, Kiaa0030, Mcmd2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM2 (Minichromosome maintenance protein 2 homolog) (Nuclear protein BM28). | |||||
|
MCM5_MOUSE
|
||||||
| θ value | 1.88829e-90 (rank : 5) | NC score | 0.947274 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
MCM7_MOUSE
|
||||||
| θ value | 5.49406e-90 (rank : 6) | NC score | 0.951435 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61881 | Gene names | Mcm7, Cdc47, Mcmd7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM7 (CDC47 homolog). | |||||
|
MCM5_HUMAN
|
||||||
| θ value | 3.56116e-89 (rank : 7) | NC score | 0.947685 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
MCM7_HUMAN
|
||||||
| θ value | 1.49618e-87 (rank : 8) | NC score | 0.950932 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33993, Q15076, Q96D34, Q96GL1 | Gene names | MCM7, CDC47, MCM2 | |||
|
Domain Architecture |
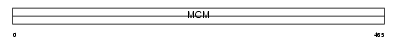
|
|||||
| Description | DNA replication licensing factor MCM7 (CDC47 homolog) (P1.1-MCM3). | |||||
|
MCM6_HUMAN
|
||||||
| θ value | 6.50502e-83 (rank : 9) | NC score | 0.941557 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14566, Q13504, Q99859 | Gene names | MCM6 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM6 (p105MCM). | |||||
|
MCM6_MOUSE
|
||||||
| θ value | 9.39325e-82 (rank : 10) | NC score | 0.941292 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97311, Q80YQ7 | Gene names | Mcm6, Mcmd6, Mis5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM6 (Mis5 homolog). | |||||
|
MCM3_HUMAN
|
||||||
| θ value | 9.0981e-77 (rank : 11) | NC score | 0.934009 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
MCM3_MOUSE
|
||||||
| θ value | 1.45253e-74 (rank : 12) | NC score | 0.930872 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P25206, Q61492 | Gene names | Mcm3, Mcmd, Mcmd3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (P1-MCM3). | |||||
|
MCM8_HUMAN
|
||||||
| θ value | 8.81223e-72 (rank : 13) | NC score | 0.932277 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UJA3, Q495R4, Q495R7, Q86US4, Q969I5 | Gene names | MCM8, C20orf154 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
MCM8_MOUSE
|
||||||
| θ value | 1.66191e-70 (rank : 14) | NC score | 0.932322 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
DYH11_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.039972 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.012327 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.009689 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.015485 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
CJ068_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.041323 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
FGD4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.014420 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FZR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.013021 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UM11, O75869, Q86U66, Q96NW8, Q9UI96, Q9ULH8, Q9UM10, Q9UNQ1, Q9Y2T8 | Gene names | FZR1, CDH1, FYR, FZR, KIAA1242 | |||
|
Domain Architecture |

|
|||||
| Description | Fizzy-related protein homolog (Fzr) (Cdh1/Hct1 homolog) (hCDH1) (CDC20-like protein 1). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.020171 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.015162 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.020081 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
ZN202_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | -0.001225 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95125, Q4JG21, Q9H1B9, Q9NSM4 | Gene names | ZNF202 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 202. | |||||
|
ECM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.020542 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.016303 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
FZR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.011683 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1K5 | Gene names | Fzr1, Fyr, Fzr | |||
|
Domain Architecture |

|
|||||
| Description | Fizzy-related protein homolog (Fzr) (Cdh1/Hct1 homolog). | |||||
|
NENF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.040586 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMX5, Q53FZ6, Q5TM90 | Gene names | NENF, SPUF | |||
|
Domain Architecture |
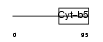
|
|||||
| Description | Neudesin precursor (Neuron-derived neurotrophic factor) (Secreted protein of unknown function) (SPUF protein). | |||||
|
CTDP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.016452 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
DYH9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.033263 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.014916 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
TAF6L_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.014817 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |
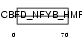
|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
ABCD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.019173 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48410, Q9QY41, Q9QZ32 | Gene names | Abcd1, Ald, Aldgh | |||
|
Domain Architecture |
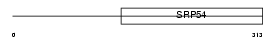
|
|||||
| Description | ATP-binding cassette sub-family D member 1 (Adrenoleukodystrophy protein) (ALDP). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.006634 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PERL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.008913 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22079, Q13408 | Gene names | LPO, SAPX | |||
|
Domain Architecture |

|
|||||
| Description | Lactoperoxidase precursor (EC 1.11.1.7) (LPO) (Salivary peroxidase) (SPO). | |||||
|
RYR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.011692 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
ABCD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.017995 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33897, Q6GTZ2 | Gene names | ABCD1, ALD | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family D member 1 (Adrenoleukodystrophy protein) (ALDP). | |||||
|
RHG18_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.013549 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.011577 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
GGN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.011883 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
SNX13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.013451 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.008543 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.001861 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.005456 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.006034 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.002559 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.004910 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TGFB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.006668 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17125 | Gene names | Tgfb3 | |||
|
Domain Architecture |
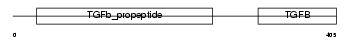
|
|||||
| Description | Transforming growth factor beta-3 precursor (TGF-beta-3). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.010374 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
ZN750_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.009672 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CAD12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.000063 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55289 | Gene names | CDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-12 precursor (Brain-cadherin) (BR-cadherin) (N-cadherin 2) (Neural type cadherin 2). | |||||
|
DCP1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.007397 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.010461 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.011540 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.005264 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.004066 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.010180 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.001038 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MCM4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MCM4_MOUSE
|
||||||
| NC score | 0.995271 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P49717, O89056 | Gene names | Mcm4, Cdc21, Mcmd4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MCM7_MOUSE
|
||||||
| NC score | 0.951435 (rank : 3) | θ value | 5.49406e-90 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61881 | Gene names | Mcm7, Cdc47, Mcmd7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM7 (CDC47 homolog). | |||||
|
MCM7_HUMAN
|
||||||
| NC score | 0.950932 (rank : 4) | θ value | 1.49618e-87 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33993, Q15076, Q96D34, Q96GL1 | Gene names | MCM7, CDC47, MCM2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM7 (CDC47 homolog) (P1.1-MCM3). | |||||
|
MCM2_HUMAN
|
||||||
| NC score | 0.948537 (rank : 5) | θ value | 5.30912e-93 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49736, Q14577, Q15023, Q8N2V1, Q969W7, Q96AE1, Q9BRM7 | Gene names | MCM2, BM28, CDCL1, KIAA0030 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM2 (Minichromosome maintenance protein 2 homolog) (Nuclear protein BM28). | |||||
|
MCM2_MOUSE
|
||||||
| NC score | 0.947974 (rank : 6) | θ value | 2.63489e-92 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97310, O08971, O89057, Q8C2R0 | Gene names | Mcm2, Bm28, Cdcl1, Kiaa0030, Mcmd2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM2 (Minichromosome maintenance protein 2 homolog) (Nuclear protein BM28). | |||||
|
MCM5_HUMAN
|
||||||
| NC score | 0.947685 (rank : 7) | θ value | 3.56116e-89 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
MCM5_MOUSE
|
||||||
| NC score | 0.947274 (rank : 8) | θ value | 1.88829e-90 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
MCM6_HUMAN
|
||||||
| NC score | 0.941557 (rank : 9) | θ value | 6.50502e-83 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14566, Q13504, Q99859 | Gene names | MCM6 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM6 (p105MCM). | |||||
|
MCM6_MOUSE
|
||||||
| NC score | 0.941292 (rank : 10) | θ value | 9.39325e-82 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97311, Q80YQ7 | Gene names | Mcm6, Mcmd6, Mis5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM6 (Mis5 homolog). | |||||
|
MCM3_HUMAN
|
||||||
| NC score | 0.934009 (rank : 11) | θ value | 9.0981e-77 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
MCM8_MOUSE
|
||||||
| NC score | 0.932322 (rank : 12) | θ value | 1.66191e-70 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
MCM8_HUMAN
|
||||||
| NC score | 0.932277 (rank : 13) | θ value | 8.81223e-72 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UJA3, Q495R4, Q495R7, Q86US4, Q969I5 | Gene names | MCM8, C20orf154 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
MCM3_MOUSE
|
||||||
| NC score | 0.930872 (rank : 14) | θ value | 1.45253e-74 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P25206, Q61492 | Gene names | Mcm3, Mcmd, Mcmd3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (P1-MCM3). | |||||
|
CJ068_HUMAN
|
||||||
| NC score | 0.041323 (rank : 15) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
NENF_HUMAN
|
||||||
| NC score | 0.040586 (rank : 16) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMX5, Q53FZ6, Q5TM90 | Gene names | NENF, SPUF | |||
|
Domain Architecture |

|
|||||
| Description | Neudesin precursor (Neuron-derived neurotrophic factor) (Secreted protein of unknown function) (SPUF protein). | |||||
|
DYH11_HUMAN
|
||||||
| NC score | 0.039972 (rank : 17) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
DYH9_HUMAN
|
||||||
| NC score | 0.033263 (rank : 18) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
ECM1_MOUSE
|
||||||
| NC score | 0.020542 (rank : 19) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.020171 (rank : 20) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.020081 (rank : 21) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
ABCD1_MOUSE
|
||||||
| NC score | 0.019173 (rank : 22) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48410, Q9QY41, Q9QZ32 | Gene names | Abcd1, Ald, Aldgh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family D member 1 (Adrenoleukodystrophy protein) (ALDP). | |||||
|
ABCD1_HUMAN
|
||||||
| NC score | 0.017995 (rank : 23) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33897, Q6GTZ2 | Gene names | ABCD1, ALD | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family D member 1 (Adrenoleukodystrophy protein) (ALDP). | |||||
|
CTDP1_MOUSE
|
||||||
| NC score | 0.016452 (rank : 24) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.016303 (rank : 25) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.015485 (rank : 26) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.015162 (rank : 27) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.014916 (rank : 28) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
TAF6L_MOUSE
|
||||||
| NC score | 0.014817 (rank : 29) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |

|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.014420 (rank : 30) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.013549 (rank : 31) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
SNX13_HUMAN
|
||||||
| NC score | 0.013451 (rank : 32) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
FZR_HUMAN
|
||||||
| NC score | 0.013021 (rank : 33) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UM11, O75869, Q86U66, Q96NW8, Q9UI96, Q9ULH8, Q9UM10, Q9UNQ1, Q9Y2T8 | Gene names | FZR1, CDH1, FYR, FZR, KIAA1242 | |||
|
Domain Architecture |

|
|||||
| Description | Fizzy-related protein homolog (Fzr) (Cdh1/Hct1 homolog) (hCDH1) (CDC20-like protein 1). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.012327 (rank : 34) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.011883 (rank : 35) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
RYR3_HUMAN
|
||||||
| NC score | 0.011692 (rank : 36) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
FZR_MOUSE
|
||||||
| NC score | 0.011683 (rank : 37) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1K5 | Gene names | Fzr1, Fyr, Fzr | |||
|
Domain Architecture |

|
|||||
| Description | Fizzy-related protein homolog (Fzr) (Cdh1/Hct1 homolog). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.011577 (rank : 38) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.011540 (rank : 39) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.010461 (rank : 40) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.010374 (rank : 41) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.010180 (rank : 42) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.009689 (rank : 43) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
ZN750_HUMAN
|
||||||
| NC score | 0.009672 (rank : 44) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
PERL_HUMAN
|
||||||
| NC score | 0.008913 (rank : 45) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22079, Q13408 | Gene names | LPO, SAPX | |||
|
Domain Architecture |

|
|||||
| Description | Lactoperoxidase precursor (EC 1.11.1.7) (LPO) (Salivary peroxidase) (SPO). | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.008543 (rank : 46) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
DCP1A_MOUSE
|
||||||
| NC score | 0.007397 (rank : 47) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
TGFB3_MOUSE
|
||||||
| NC score | 0.006668 (rank : 48) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17125 | Gene names | Tgfb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor beta-3 precursor (TGF-beta-3). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.006634 (rank : 49) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.006034 (rank : 50) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.005456 (rank : 51) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.005264 (rank : 52) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.004910 (rank : 53) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.004066 (rank : 54) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.002559 (rank : 55) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.001861 (rank : 56) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.001038 (rank : 57) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CAD12_HUMAN
|
||||||
| NC score | 0.000063 (rank : 58) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55289 | Gene names | CDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-12 precursor (Brain-cadherin) (BR-cadherin) (N-cadherin 2) (Neural type cadherin 2). | |||||
|
ZN202_HUMAN
|
||||||
| NC score | -0.001225 (rank : 59) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95125, Q4JG21, Q9H1B9, Q9NSM4 | Gene names | ZNF202 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 202. | |||||