Please be patient as the page loads
|
RYR3_HUMAN
|
||||||
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RYR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.961454 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.975845 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
RYR3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
ITPR1_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 4) | NC score | 0.562619 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 5) | NC score | 0.558716 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
ITPR3_MOUSE
|
||||||
| θ value | 4.14116e-29 (rank : 6) | NC score | 0.546790 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR2_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 7) | NC score | 0.541929 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ITPR2_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 8) | NC score | 0.541309 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
ITPR3_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 9) | NC score | 0.542713 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
RN123_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 10) | NC score | 0.367348 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5XPI4, Q5I022, Q6PFW4, Q71RH0, Q8IW18, Q9H0M8, Q9H5L8, Q9H9T2 | Gene names | RNF123, KPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
RN123_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 11) | NC score | 0.366471 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5XPI3, Q6PGE0 | Gene names | Rnf123, Kpc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
ASH2L_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 12) | NC score | 0.268203 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBL3, O60659, O60660, Q96B62 | Gene names | ASH2L | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
ASH2L_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 13) | NC score | 0.267440 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
RANB9_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 14) | NC score | 0.223327 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
RANB9_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 15) | NC score | 0.236218 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 16) | NC score | 0.111246 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 17) | NC score | 0.108731 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
MLRN_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 18) | NC score | 0.088899 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
CALM_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.074008 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.074008 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.020580 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.073369 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
ADIP_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.048259 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.072867 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
OSBL3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 25) | NC score | 0.025363 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.076580 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MLRM_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.085797 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |
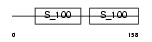
|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
MLRN_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.085432 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |
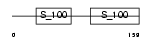
|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.070284 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
DJC12_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.033149 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
BIRC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.022079 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
GPT_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.050275 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3H5, O15216, Q86WV9, Q9BWE6 | Gene names | DPAGT1, DPAGT2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--dolichyl-phosphate N- acetylglucosaminephosphotransferase (EC 2.7.8.15) (GPT) (G1PT) (N- acetylglucosamine-1-phosphate transferase) (GlcNAc-1-P transferase). | |||||
|
MLR5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.095552 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |
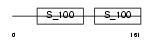
|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.038606 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
EFCB2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.071555 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
PINX1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.039141 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
EFHB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.056042 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
ADIP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.035553 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2D8, Q6P2P8, Q6ULS1, Q7L168, Q9UIX0 | Gene names | SSX2IP, KIAA0923 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein) (SSX2-interacting protein). | |||||
|
MLRV_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.092901 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |
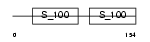
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
CABP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.064084 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |
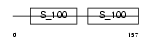
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
NALDL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.019365 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQQ1, O43176 | Gene names | NAALADL1, NAALADASEL, NAALADL | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L) (Ileal dipeptidylpeptidase) (100 kDa ileum brush border membrane protein) (I100). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.018686 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
STAM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.012469 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
AR13B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.019210 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
CABP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.062954 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
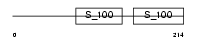
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
DJBP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.061927 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
MLRS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.088656 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |
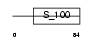
|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
SPRY4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.029622 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WW59 | Gene names | SPRYD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPRY domain-containing protein 4. | |||||
|
SPRY4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.045898 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91WK1, Q8BUS6, Q9D651 | Gene names | Spryd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPRY domain-containing protein 4. | |||||
|
CABP3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.068856 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |
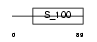
|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.066124 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
FBX40_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.016780 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62932 | Gene names | Fbxo40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40. | |||||
|
HAPIP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.016328 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
MLRA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.083838 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.083940 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRV_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.084656 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.024272 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
SOMA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.037150 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01241, Q14405, Q16631, Q5EB53, Q9HBZ1, Q9UMJ7, Q9UNL5 | Gene names | GH1 | |||
|
Domain Architecture |
|
|||||
| Description | Somatotropin precursor (Growth hormone) (GH) (GH-N) (Pituitary growth hormone) (Growth hormone 1). | |||||
|
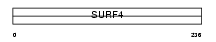
SURF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.033022 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15260, O60923, Q9UNZ0, Q9UNZ1 | Gene names | SURF4, SURF-4 | |||
|
Domain Architecture |
|
|||||
| Description | Surfeit locus protein 4. | |||||
|
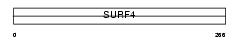
SURF4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.033019 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64310 | Gene names | Surf4, Surf-4 | |||
|
Domain Architecture |
|
|||||
| Description | Surfeit locus protein 4. | |||||
|
TRIM9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.040778 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C026, Q92557, Q96D24, Q96NI4, Q9C025, Q9C027 | Gene names | TRIM9, KIAA0282, RNF91 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9 (RING finger protein 91). | |||||
|
XPOT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.025637 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43592, O43784, Q8WUG2, Q9BVS7 | Gene names | XPOT | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-T (tRNA exportin) (Exportin(tRNA)). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.069753 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
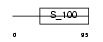
CABP7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.066510 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
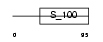
CABP7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.066510 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.012060 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CF152_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.016471 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.015252 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
EFHB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.051118 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
GFRA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.012669 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56159, O15507, O43912 | Gene names | GFRA1, GDNFRA, RETL1, TRNR1 | |||
|
Domain Architecture |
|
|||||
| Description | GDNF family receptor alpha-1 precursor (GFR-alpha-1) (GDNF receptor alpha) (GDNFR-alpha) (TGF-beta-related neurotrophic factor receptor 1) (RET ligand 1). | |||||
|
GPT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.035879 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42867, Q921W5 | Gene names | Dpagt1, Dpagt2, Gnpta | |||
|
Domain Architecture |
|
|||||
| Description | UDP-N-acetylglucosamine--dolichyl-phosphate N- acetylglucosaminephosphotransferase (EC 2.7.8.15) (GPT) (G1PT) (N- acetylglucosamine-1-phosphate transferase) (GlcNAc-1-P transferase). | |||||
|
LIPR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.013246 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17892 | Gene names | Pnliprp2, Plrp2 | |||
|
Domain Architecture |
|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3) (Cytotoxic T-lymphocyte lipase). | |||||
|
MCM4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.011692 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.008468 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
PPME1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.026873 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y570, Q9NVT5, Q9UI18 | Gene names | PPME1, PME1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.009694 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.011811 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.021607 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TRIM9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.039991 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C7M3, Q6ZQE5, Q80WT6, Q8C7Z4, Q8CC32, Q8CEG2, Q99PQ3 | Gene names | Trim9, Kiaa0282 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9. | |||||
|
UBCP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.034516 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVY7, Q96DK5 | Gene names | UBLCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
UBCP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.034513 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGR9, Q52L62, Q5XJH1, Q6PH96, Q99LT3 | Gene names | Ublcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
UBR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.016491 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70481, Q5BKR8, Q792M3, Q8BN40, Q8C5K3 | Gene names | Ubr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.010984 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CADH6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.001862 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97326, P70393 | Gene names | Cdh6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
CF010_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.019936 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
DMXL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.017179 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
DPOD3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.017073 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
KR134_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.017888 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI77 | Gene names | KRTAP13-4, KAP13.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-4. | |||||
|
LRCH1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.005395 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2L9, Q7Z5F6, Q7Z5F7 | Gene names | LRCH1, CHDC1, KIAA1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1) (Neuronal protein 81) (NP81). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.001581 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
PPME1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.025019 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVQ5, Q3U392, Q3UJX8, Q3UKE0, Q8K311, Q91YR8, Q9CSP7 | Gene names | Ppme1, Pme1 | |||
|
Domain Architecture |
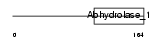
|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.012139 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.010783 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
ALBU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.009376 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07724, Q61802 | Gene names | Alb, Alb-1, Alb1 | |||
|
Domain Architecture |

|
|||||
| Description | Serum albumin precursor. | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.014696 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.016781 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
NALDL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.014467 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M758 | Gene names | Naaladl1, Naaladasel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L). | |||||
|
REST_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.009310 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.006627 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
UBR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.014845 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWV7, O60708, O75492, Q68DN9, Q8IWY6, Q96JY4 | Gene names | UBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
UNC5B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.013206 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
ANC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.011479 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTZ4, Q3TGA7, Q80UJ6, Q80W43, Q8BWW7, Q8C0F7, Q9CRX0, Q9CSL1, Q9JJB8 | Gene names | Anapc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
CBWD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.013338 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEH6, Q7TSA8, Q8K399 | Gene names | Cbwd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COBW domain-containing protein 1. | |||||
|
FA40B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.008764 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C9H6, Q3TQB7, Q80TI6, Q8C7A2 | Gene names | Fam40b, Kiaa1170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM40B. | |||||
|
GEMI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.017931 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
HIC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | -0.002498 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLZ6, Q3U030, Q3ULP4, Q5K036, Q8BSZ9, Q8C3T5 | Gene names | Hic2, Kiaa1020 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.033179 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
JIP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.018730 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
K0494_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.018738 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75071 | Gene names | KIAA0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
LIMK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | -0.000695 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53671, Q7KZ80, Q96E10, Q99464 | Gene names | LIMK2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain kinase 2 (EC 2.7.11.1) (LIMK-2). | |||||
|
LIPR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.010497 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54317 | Gene names | PNLIPRP2, PLRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.009490 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.011310 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PITM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.006059 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PTN22_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.003022 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
STAG3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.006989 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJ98, Q8NDP3 | Gene names | STAG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.009631 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.006009 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UNC5B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.013162 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
CCP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.007894 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHG6, Q91WQ4, Q9JK50, Q9JK51, Q9JKK2, Q9JKK3 | Gene names | Dscr1 | |||
|
Domain Architecture |
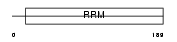
|
|||||
| Description | Calcipressin-1 (Down syndrome critical region protein 1 homolog) (Myocyte-enriched calcineurin-interacting protein 1) (MCIP1). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.011244 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.051785 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
DMN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.004212 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.005215 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.006947 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MCM4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.008864 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49717, O89056 | Gene names | Mcm4, Cdc21, Mcmd4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MERTK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | -0.002177 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60805, Q62194 | Gene names | Mertk, Mer | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase MER precursor (EC 2.7.10.1) (C- mer) (Receptor tyrosine kinase MerTK). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.006921 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
O10AD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | -0.000789 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGE0 | Gene names | OR10AD1 | |||
|
Domain Architecture |
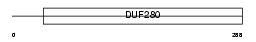
|
|||||
| Description | Olfactory receptor 10AD1. | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.007121 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.014683 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TTF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.009939 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
TULP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.006901 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00295 | Gene names | TULP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 2 (Tubby-like protein 2). | |||||
|
VDP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.009169 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
CABP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.052447 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
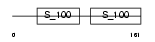
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.053051 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
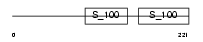
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CALL6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.061993 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.050473 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CALN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.050371 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |
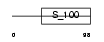
|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.050374 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |
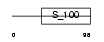
|
|||||
| Description | Calneuron-1. | |||||
|
HNRL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.095807 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.095918 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
RYR3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.975845 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.961454 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
ITPR1_HUMAN
|
||||||
| NC score | 0.562619 (rank : 4) | θ value | 6.59618e-35 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.558716 (rank : 5) | θ value | 2.77131e-33 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
ITPR3_MOUSE
|
||||||
| NC score | 0.546790 (rank : 6) | θ value | 4.14116e-29 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR3_HUMAN
|
||||||
| NC score | 0.542713 (rank : 7) | θ value | 1.33217e-27 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR2_MOUSE
|
||||||
| NC score | 0.541929 (rank : 8) | θ value | 1.02001e-27 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ITPR2_HUMAN
|
||||||
| NC score | 0.541309 (rank : 9) | θ value | 1.33217e-27 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
RN123_HUMAN
|
||||||
| NC score | 0.367348 (rank : 10) | θ value | 4.91457e-14 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5XPI4, Q5I022, Q6PFW4, Q71RH0, Q8IW18, Q9H0M8, Q9H5L8, Q9H9T2 | Gene names | RNF123, KPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
RN123_MOUSE
|
||||||
| NC score | 0.366471 (rank : 11) | θ value | 4.91457e-14 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5XPI3, Q6PGE0 | Gene names | Rnf123, Kpc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
ASH2L_HUMAN
|
||||||
| NC score | 0.268203 (rank : 12) | θ value | 2.88788e-06 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBL3, O60659, O60660, Q96B62 | Gene names | ASH2L | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
ASH2L_MOUSE
|
||||||
| NC score | 0.267440 (rank : 13) | θ value | 2.88788e-06 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
RANB9_MOUSE
|
||||||
| NC score | 0.236218 (rank : 14) | θ value | 0.000158464 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
RANB9_HUMAN
|
||||||
| NC score | 0.223327 (rank : 15) | θ value | 0.000158464 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.111246 (rank : 16) | θ value | 0.00035302 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.108731 (rank : 17) | θ value | 0.000602161 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.095918 (rank : 18) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
HNRL1_HUMAN
|
||||||
| NC score | 0.095807 (rank : 19) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
MLR5_HUMAN
|
||||||
| NC score | 0.095552 (rank : 20) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |

|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
MLRV_HUMAN
|
||||||
| NC score | 0.092901 (rank : 21) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRN_MOUSE
|
||||||
| NC score | 0.088899 (rank : 22) | θ value | 0.0193708 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MLRS_MOUSE
|
||||||
| NC score | 0.088656 (rank : 23) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
MLRM_HUMAN
|
||||||
| NC score | 0.085797 (rank : 24) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
MLRN_HUMAN
|
||||||
| NC score | 0.085432 (rank : 25) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MLRV_MOUSE
|
||||||
| NC score | 0.084656 (rank : 26) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRA_MOUSE
|
||||||
| NC score | 0.083940 (rank : 27) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRA_HUMAN
|
||||||
| NC score | 0.083838 (rank : 28) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.076580 (rank : 29) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.074008 (rank : 30) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.074008 (rank : 31) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.073369 (rank : 32) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.072867 (rank : 33) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
EFCB2_MOUSE
|
||||||
| NC score | 0.071555 (rank : 34) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.070284 (rank : 35) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.069753 (rank : 36) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP3_HUMAN
|
||||||
| NC score | 0.068856 (rank : 37) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP7_HUMAN
|
||||||
| NC score | 0.066510 (rank : 38) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| NC score | 0.066510 (rank : 39) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.066124 (rank : 40) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP2_MOUSE
|
||||||
| NC score | 0.064084 (rank : 41) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP2_HUMAN
|
||||||
| NC score | 0.062954 (rank : 42) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
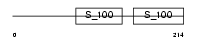
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CALL6_HUMAN
|
||||||
| NC score | 0.061993 (rank : 43) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
DJBP_HUMAN
|
||||||
| NC score | 0.061927 (rank : 44) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
EFHB_HUMAN
|
||||||
| NC score | 0.056042 (rank : 45) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.053051 (rank : 46) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.052447 (rank : 47) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.051785 (rank : 48) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
EFHB_MOUSE
|
||||||
| NC score | 0.051118 (rank : 49) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.050473 (rank : 50) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CALN1_MOUSE
|
||||||
| NC score | 0.050374 (rank : 51) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1. | |||||
|
CALN1_HUMAN
|
||||||
| NC score | 0.050371 (rank : 52) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
GPT_HUMAN
|
||||||
| NC score | 0.050275 (rank : 53) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3H5, O15216, Q86WV9, Q9BWE6 | Gene names | DPAGT1, DPAGT2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--dolichyl-phosphate N- acetylglucosaminephosphotransferase (EC 2.7.8.15) (GPT) (G1PT) (N- acetylglucosamine-1-phosphate transferase) (GlcNAc-1-P transferase). | |||||
|
ADIP_MOUSE
|
||||||
| NC score | 0.048259 (rank : 54) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
SPRY4_MOUSE
|
||||||
| NC score | 0.045898 (rank : 55) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91WK1, Q8BUS6, Q9D651 | Gene names | Spryd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPRY domain-containing protein 4. | |||||
|
TRIM9_HUMAN
|
||||||
| NC score | 0.040778 (rank : 56) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C026, Q92557, Q96D24, Q96NI4, Q9C025, Q9C027 | Gene names | TRIM9, KIAA0282, RNF91 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9 (RING finger protein 91). | |||||
|
TRIM9_MOUSE
|
||||||
| NC score | 0.039991 (rank : 57) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C7M3, Q6ZQE5, Q80WT6, Q8C7Z4, Q8CC32, Q8CEG2, Q99PQ3 | Gene names | Trim9, Kiaa0282 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9. | |||||
|
PINX1_HUMAN
|
||||||
| NC score | 0.039141 (rank : 58) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.038606 (rank : 59) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
SOMA_HUMAN
|
||||||
| NC score | 0.037150 (rank : 60) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01241, Q14405, Q16631, Q5EB53, Q9HBZ1, Q9UMJ7, Q9UNL5 | Gene names | GH1 | |||
|
Domain Architecture |

|
|||||
| Description | Somatotropin precursor (Growth hormone) (GH) (GH-N) (Pituitary growth hormone) (Growth hormone 1). | |||||
|
GPT_MOUSE
|
||||||
| NC score | 0.035879 (rank : 61) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42867, Q921W5 | Gene names | Dpagt1, Dpagt2, Gnpta | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--dolichyl-phosphate N- acetylglucosaminephosphotransferase (EC 2.7.8.15) (GPT) (G1PT) (N- acetylglucosamine-1-phosphate transferase) (GlcNAc-1-P transferase). | |||||
|
ADIP_HUMAN
|
||||||
| NC score | 0.035553 (rank : 62) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2D8, Q6P2P8, Q6ULS1, Q7L168, Q9UIX0 | Gene names | SSX2IP, KIAA0923 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein) (SSX2-interacting protein). | |||||
|
UBCP1_HUMAN
|
||||||
| NC score | 0.034516 (rank : 63) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVY7, Q96DK5 | Gene names | UBLCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
UBCP1_MOUSE
|
||||||
| NC score | 0.034513 (rank : 64) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGR9, Q52L62, Q5XJH1, Q6PH96, Q99LT3 | Gene names | Ublcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.033179 (rank : 65) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DJC12_HUMAN
|
||||||
| NC score | 0.033149 (rank : 66) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
SURF4_HUMAN
|
||||||
| NC score | 0.033022 (rank : 67) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15260, O60923, Q9UNZ0, Q9UNZ1 | Gene names | SURF4, SURF-4 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 4. | |||||
|
SURF4_MOUSE
|
||||||
| NC score | 0.033019 (rank : 68) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64310 | Gene names | Surf4, Surf-4 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 4. | |||||
|
SPRY4_HUMAN
|
||||||
| NC score | 0.029622 (rank : 69) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WW59 | Gene names | SPRYD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPRY domain-containing protein 4. | |||||
|
PPME1_HUMAN
|
||||||
| NC score | 0.026873 (rank : 70) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y570, Q9NVT5, Q9UI18 | Gene names | PPME1, PME1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
XPOT_HUMAN
|
||||||
| NC score | 0.025637 (rank : 71) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43592, O43784, Q8WUG2, Q9BVS7 | Gene names | XPOT | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-T (tRNA exportin) (Exportin(tRNA)). | |||||
|
OSBL3_MOUSE
|
||||||
| NC score | 0.025363 (rank : 72) | θ value | 0.0431538 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
PPME1_MOUSE
|
||||||
| NC score | 0.025019 (rank : 73) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVQ5, Q3U392, Q3UJX8, Q3UKE0, Q8K311, Q91YR8, Q9CSP7 | Gene names | Ppme1, Pme1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.024272 (rank : 74) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
BIRC2_HUMAN
|
||||||
| NC score | 0.022079 (rank : 75) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.021607 (rank : 76) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.020580 (rank : 77) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.019936 (rank : 78) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
NALDL_HUMAN
|
||||||
| NC score | 0.019365 (rank : 79) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQQ1, O43176 | Gene names | NAALADL1, NAALADASEL, NAALADL | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L) (Ileal dipeptidylpeptidase) (100 kDa ileum brush border membrane protein) (I100). | |||||
|
AR13B_HUMAN
|
||||||
| NC score | 0.019210 (rank : 80) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
K0494_HUMAN
|
||||||
| NC score | 0.018738 (rank : 81) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75071 | Gene names | KIAA0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
JIP3_MOUSE
|
||||||
| NC score | 0.018730 (rank : 82) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.018686 (rank : 83) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
GEMI_MOUSE
|
||||||
| NC score | 0.017931 (rank : 84) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
KR134_HUMAN
|
||||||
| NC score | 0.017888 (rank : 85) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI77 | Gene names | KRTAP13-4, KAP13.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-4. | |||||
|
DMXL2_HUMAN
|
||||||
| NC score | 0.017179 (rank : 86) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
DPOD3_MOUSE
|
||||||
| NC score | 0.017073 (rank : 87) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.016781 (rank : 88) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
FBX40_MOUSE
|
||||||
| NC score | 0.016780 (rank : 89) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62932 | Gene names | Fbxo40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40. | |||||
|
UBR1_MOUSE
|
||||||
| NC score | 0.016491 (rank : 90) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70481, Q5BKR8, Q792M3, Q8BN40, Q8C5K3 | Gene names | Ubr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.016471 (rank : 91) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
HAPIP_HUMAN
|
||||||
| NC score | 0.016328 (rank : 92) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.015252 (rank : 93) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
UBR1_HUMAN
|
||||||
| NC score | 0.014845 (rank : 94) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWV7, O60708, O75492, Q68DN9, Q8IWY6, Q96JY4 | Gene names | UBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.014696 (rank : 95) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.014683 (rank : 96) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
NALDL_MOUSE
|
||||||
| NC score | 0.014467 (rank : 97) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M758 | Gene names | Naaladl1, Naaladasel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L). | |||||
|
CBWD1_MOUSE
|
||||||
| NC score | 0.013338 (rank : 98) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEH6, Q7TSA8, Q8K399 | Gene names | Cbwd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COBW domain-containing protein 1. | |||||
|
LIPR2_MOUSE
|
||||||
| NC score | 0.013246 (rank : 99) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17892 | Gene names | Pnliprp2, Plrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3) (Cytotoxic T-lymphocyte lipase). | |||||
|
UNC5B_HUMAN
|
||||||
| NC score | 0.013206 (rank : 100) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
UNC5B_MOUSE
|
||||||
| NC score | 0.013162 (rank : 101) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
GFRA1_HUMAN
|
||||||
| NC score | 0.012669 (rank : 102) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56159, O15507, O43912 | Gene names | GFRA1, GDNFRA, RETL1, TRNR1 | |||
|
Domain Architecture |

|
|||||
| Description | GDNF family receptor alpha-1 precursor (GFR-alpha-1) (GDNF receptor alpha) (GDNFR-alpha) (TGF-beta-related neurotrophic factor receptor 1) (RET ligand 1). | |||||
|
STAM2_HUMAN
|
||||||
| NC score | 0.012469 (rank : 103) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.012139 (rank : 104) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.012060 (rank : 105) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.011811 (rank : 106) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
MCM4_HUMAN
|
||||||
| NC score | 0.011692 (rank : 107) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
ANC5_MOUSE
|
||||||
| NC score | 0.011479 (rank : 108) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTZ4, Q3TGA7, Q80UJ6, Q80W43, Q8BWW7, Q8C0F7, Q9CRX0, Q9CSL1, Q9JJB8 | Gene names | Anapc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.011310 (rank : 109) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.011244 (rank : 110) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.010984 (rank : 111) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.010783 (rank : 112) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
LIPR2_HUMAN
|
||||||
| NC score | 0.010497 (rank : 113) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54317 | Gene names | PNLIPRP2, PLRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3). | |||||
|
TTF1_MOUSE
|
||||||
| NC score | 0.009939 (rank : 114) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.009694 (rank : 115) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.009631 (rank : 116) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.009490 (rank : 117) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ALBU_MOUSE
|
||||||
| NC score | 0.009376 (rank : 118) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07724, Q61802 | Gene names | Alb, Alb-1, Alb1 | |||
|
Domain Architecture |

|
|||||
| Description | Serum albumin precursor. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.009310 (rank : 119) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
VDP_HUMAN
|
||||||
| NC score | 0.009169 (rank : 120) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
MCM4_MOUSE
|
||||||
| NC score | 0.008864 (rank : 121) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49717, O89056 | Gene names | Mcm4, Cdc21, Mcmd4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
FA40B_MOUSE
|
||||||
| NC score | 0.008764 (rank : 122) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C9H6, Q3TQB7, Q80TI6, Q8C7A2 | Gene names | Fam40b, Kiaa1170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM40B. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.008468 (rank : 123) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
CCP1_MOUSE
|
||||||
| NC score | 0.007894 (rank : 124) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHG6, Q91WQ4, Q9JK50, Q9JK51, Q9JKK2, Q9JKK3 | Gene names | Dscr1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcipressin-1 (Down syndrome critical region protein 1 homolog) (Myocyte-enriched calcineurin-interacting protein 1) (MCIP1). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.007121 (rank : 125) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
STAG3_HUMAN
|
||||||
| NC score | 0.006989 (rank : 126) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJ98, Q8NDP3 | Gene names | STAG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.006947 (rank : 127) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.006921 (rank : 128) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
TULP2_HUMAN
|
||||||
| NC score | 0.006901 (rank : 129) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00295 | Gene names | TULP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 2 (Tubby-like protein 2). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.006627 (rank : 130) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PITM2_MOUSE
|
||||||
| NC score | 0.006059 (rank : 131) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.006009 (rank : 132) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
LRCH1_HUMAN
|
||||||
| NC score | 0.005395 (rank : 133) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2L9, Q7Z5F6, Q7Z5F7 | Gene names | LRCH1, CHDC1, KIAA1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1) (Neuronal protein 81) (NP81). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.005215 (rank : 134) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
DMN_HUMAN
|
||||||
| NC score | 0.004212 (rank : 135) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
PTN22_HUMAN
|
||||||
| NC score | 0.003022 (rank : 136) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
CADH6_MOUSE
|
||||||
| NC score | 0.001862 (rank : 137) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97326, P70393 | Gene names | Cdh6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.001581 (rank : 138) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
LIMK2_HUMAN
|
||||||
| NC score | -0.000695 (rank : 139) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53671, Q7KZ80, Q96E10, Q99464 | Gene names | LIMK2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain kinase 2 (EC 2.7.11.1) (LIMK-2). | |||||
|
O10AD_HUMAN
|
||||||
| NC score | -0.000789 (rank : 140) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGE0 | Gene names | OR10AD1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 10AD1. | |||||
|
MERTK_MOUSE
|
||||||
| NC score | -0.002177 (rank : 141) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60805, Q62194 | Gene names | Mertk, Mer | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase MER precursor (EC 2.7.10.1) (C- mer) (Receptor tyrosine kinase MerTK). | |||||
|
HIC2_MOUSE
|
||||||
| NC score | -0.002498 (rank : 142) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLZ6, Q3U030, Q3ULP4, Q5K036, Q8BSZ9, Q8C3T5 | Gene names | Hic2, Kiaa1020 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2). | |||||