Please be patient as the page loads
|
ITPR3_HUMAN
|
||||||
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ITPR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989276 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.988722 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
ITPR2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.990631 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
ITPR2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.991022 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ITPR3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.997177 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 3.50572e-28 (rank : 7) | NC score | 0.532386 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 4.57862e-28 (rank : 8) | NC score | 0.536980 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
RYR3_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 9) | NC score | 0.542713 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 10) | NC score | 0.053057 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
SYTL4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.017333 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.046815 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.016479 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.039053 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.039116 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.038337 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.040097 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.032552 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PAIP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.047374 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VE62, Q9WUC9 | Gene names | Paip1 | |||
|
Domain Architecture |
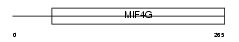
|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.021875 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.035378 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.026460 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.035437 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
VIME_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.020795 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P08670, Q15867, Q15868, Q15869, Q6LER9, Q8N850, Q96ML2, Q9NTM3 | Gene names | VIM | |||
|
Domain Architecture |

|
|||||
| Description | Vimentin. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.033213 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
EP15_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.047385 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
GP158_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.032764 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.027990 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.020748 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.019584 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
WDHD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.020218 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.014916 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.036716 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.034389 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.019939 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.025073 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.044214 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
PAIP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.037047 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.050991 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.035716 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
KINH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.025388 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.029969 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.015789 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.019330 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
GBGT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.030111 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14610 | Gene names | GNGT2, GNG8, GNG9, GNGT8 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-T2 subunit precursor (G gamma-C) (G-gamma-8) (G-gamma 9). | |||||
|
GCP6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.029958 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.037053 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.011673 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
ABCAD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.007887 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
CF010_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.020078 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.032326 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.014630 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.024214 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.033429 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
INT2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.020409 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UK8, Q5SXZ5, Q5SXZ6, Q6PCY7, Q6ZPU6, Q8CB15, Q9CSE0 | Gene names | Ints2, Kiaa1287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 2 (Int2). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.032589 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RRP44_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.027200 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.020606 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
APHC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.029786 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NUN7 | Gene names | PHCA, APHC | |||
|
Domain Architecture |
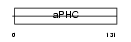
|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase) (Alkaline dihydroceramidase SB89). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.037532 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CJ068_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.033634 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
CN145_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.034519 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
ERF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.018925 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62495, P46055, Q5M7Z7 | Gene names | ETF1, ERF1, RF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic peptide chain release factor subunit 1 (eRF1) (Eukaryotic release factor 1) (TB3-1) (Cl1 protein). | |||||
|
ERF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.018925 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BWY3, Q3TPZ6, Q91VH9 | Gene names | Etf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic peptide chain release factor subunit 1 (eRF1) (Eukaryotic release factor 1). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.032429 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.026899 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.027143 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.029027 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.028417 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYO5B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.022736 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
NPAL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.019962 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGN5, Q6PG85, Q9D2Y2 | Gene names | Npal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NIPA-like protein 3. | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.025820 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
SYTL4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.010921 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
TXD13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.012882 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |
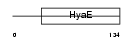
|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.000760 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.031541 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.031023 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.005064 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
BCAS4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.012135 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDM0, Q5TD52, Q5TD53, Q5TD54, Q5U5K7, Q5XKE8, Q8IXI7, Q8NEZ6, Q8TDL9, Q9NX13, Q9Y511 | Gene names | BCAS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 4. | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.009645 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.009800 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
COVA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.026961 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |
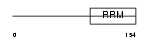
|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.010418 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.030456 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.001576 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
GTR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.006517 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11168 | Gene names | SLC2A2, GLUT2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 2 (Glucose transporter type 2, liver). | |||||
|
HIPK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.000921 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86Z02, O75125, Q8IYD7, Q8NDN5, Q8NEB6, Q8TBZ1 | Gene names | HIPK1, KIAA0630 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1). | |||||
|
KGP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.002976 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.016125 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.018805 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.027801 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.033225 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.005115 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.027246 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NPAL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.016357 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P499, Q6MZT9, Q9BVE6 | Gene names | NPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NIPA-like protein 3. | |||||
|
NUF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.036304 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
OLR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.024196 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EQ09 | Gene names | Olr1, Lox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.040953 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
SL9A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.008144 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19634 | Gene names | SLC9A1, APNH1, NHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 1 (Na(+)/H(+) exchanger 1) (NHE-1) (Solute carrier family 9 member 1) (Na(+)/H(+) antiporter, amiloride- sensitive) (APNH). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.013345 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.021982 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
AOF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.005236 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.027900 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.027690 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.026655 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.031124 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
DZIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.019203 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
IGB1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.007783 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ29 | Gene names | Igbp1b | |||
|
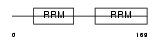
Domain Architecture |
|
|||||
| Description | Immunoglobulin-binding protein 1b (CD79a-binding protein 1b) (Alpha 4- b protein) (Alpha4-b). | |||||
|
KKCC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.000454 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VBY2, Q9R054 | Gene names | Camkk1, Camkk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.025844 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.026606 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
PANK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.004965 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.028435 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
RBMS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.005321 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
ITPR3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR3_MOUSE
|
||||||
| NC score | 0.997177 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR2_MOUSE
|
||||||
| NC score | 0.991022 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ITPR2_HUMAN
|
||||||
| NC score | 0.990631 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
ITPR1_HUMAN
|
||||||
| NC score | 0.989276 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.988722 (rank : 6) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
RYR3_HUMAN
|
||||||
| NC score | 0.542713 (rank : 7) | θ value | 1.33217e-27 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.536980 (rank : 8) | θ value | 4.57862e-28 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.532386 (rank : 9) | θ value | 3.50572e-28 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.053057 (rank : 10) | θ value | 0.0148317 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.050991 (rank : 11) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.047385 (rank : 12) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
PAIP1_MOUSE
|
||||||
| NC score | 0.047374 (rank : 13) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VE62, Q9WUC9 | Gene names | Paip1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.046815 (rank : 14) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.044214 (rank : 15) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.040953 (rank : 16) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.040097 (rank : 17) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.039116 (rank : 18) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.039053 (rank : 19) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.038337 (rank : 20) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.037532 (rank : 21) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.037053 (rank : 22) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
PAIP1_HUMAN
|
||||||
| NC score | 0.037047 (rank : 23) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.036716 (rank : 24) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.036304 (rank : 25) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.035716 (rank : 26) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.035437 (rank : 27) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.035378 (rank : 28) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.034519 (rank : 29) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.034389 (rank : 30) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CJ068_HUMAN
|
||||||
| NC score | 0.033634 (rank : 31) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.033429 (rank : 32) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.033225 (rank : 33) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.033213 (rank : 34) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
GP158_MOUSE
|
||||||
| NC score | 0.032764 (rank : 35) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.032589 (rank : 36) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.032552 (rank : 37) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.032429 (rank : 38) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.032326 (rank : 39) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.031541 (rank : 40) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.031124 (rank : 41) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.031023 (rank : 42) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.030456 (rank : 43) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
GBGT2_HUMAN
|
||||||
| NC score | 0.030111 (rank : 44) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14610 | Gene names | GNGT2, GNG8, GNG9, GNGT8 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-T2 subunit precursor (G gamma-C) (G-gamma-8) (G-gamma 9). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.029969 (rank : 45) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
GCP6_HUMAN
|
||||||
| NC score | 0.029958 (rank : 46) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
APHC_HUMAN
|
||||||
| NC score | 0.029786 (rank : 47) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NUN7 | Gene names | PHCA, APHC | |||
|
Domain Architecture |

|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase) (Alkaline dihydroceramidase SB89). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.029027 (rank : 48) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.028435 (rank : 49) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.028417 (rank : 50) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.027990 (rank : 51) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.027900 (rank : 52) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.027801 (rank : 53) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.027690 (rank : 54) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.027246 (rank : 55) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
RRP44_HUMAN
|
||||||
| NC score | 0.027200 (rank : 56) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.027143 (rank : 57) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
COVA1_HUMAN
|
||||||
| NC score | 0.026961 (rank : 58) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.026899 (rank : 59) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.026655 (rank : 60) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.026606 (rank : 61) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.026460 (rank : 62) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.025844 (rank : 63) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.025820 (rank : 64) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.025388 (rank : 65) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.025073 (rank : 66) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.024214 (rank : 67) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
OLR1_MOUSE
|
||||||
| NC score | 0.024196 (rank : 68) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EQ09 | Gene names | Olr1, Lox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
MYO5B_HUMAN
|
||||||
| NC score | 0.022736 (rank : 69) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.021982 (rank : 70) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.021875 (rank : 71) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
VIME_HUMAN
|
||||||
| NC score | 0.020795 (rank : 72) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P08670, Q15867, Q15868, Q15869, Q6LER9, Q8N850, Q96ML2, Q9NTM3 | Gene names | VIM | |||
|
Domain Architecture |

|
|||||
| Description | Vimentin. | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.020748 (rank : 73) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.020606 (rank : 74) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
INT2_MOUSE
|
||||||
| NC score | 0.020409 (rank : 75) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UK8, Q5SXZ5, Q5SXZ6, Q6PCY7, Q6ZPU6, Q8CB15, Q9CSE0 | Gene names | Ints2, Kiaa1287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 2 (Int2). | |||||
|
WDHD1_HUMAN
|
||||||
| NC score | 0.020218 (rank : 76) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.020078 (rank : 77) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
NPAL3_MOUSE
|
||||||
| NC score | 0.019962 (rank : 78) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGN5, Q6PG85, Q9D2Y2 | Gene names | Npal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NIPA-like protein 3. | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.019939 (rank : 79) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.019584 (rank : 80) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.019330 (rank : 81) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
DZIP1_MOUSE
|
||||||
| NC score | 0.019203 (rank : 82) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
ERF1_HUMAN
|
||||||
| NC score | 0.018925 (rank : 83) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62495, P46055, Q5M7Z7 | Gene names | ETF1, ERF1, RF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic peptide chain release factor subunit 1 (eRF1) (Eukaryotic release factor 1) (TB3-1) (Cl1 protein). | |||||
|
ERF1_MOUSE
|
||||||
| NC score | 0.018925 (rank : 84) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BWY3, Q3TPZ6, Q91VH9 | Gene names | Etf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic peptide chain release factor subunit 1 (eRF1) (Eukaryotic release factor 1). | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.018805 (rank : 85) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
SYTL4_MOUSE
|
||||||
| NC score | 0.017333 (rank : 86) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.016479 (rank : 87) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
NPAL3_HUMAN
|
||||||
| NC score | 0.016357 (rank : 88) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P499, Q6MZT9, Q9BVE6 | Gene names | NPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NIPA-like protein 3. | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.016125 (rank : 89) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.015789 (rank : 90) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.014916 (rank : 91) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.014630 (rank : 92) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.013345 (rank : 93) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
TXD13_HUMAN
|
||||||
| NC score | 0.012882 (rank : 94) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
BCAS4_HUMAN
|
||||||
| NC score | 0.012135 (rank : 95) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDM0, Q5TD52, Q5TD53, Q5TD54, Q5U5K7, Q5XKE8, Q8IXI7, Q8NEZ6, Q8TDL9, Q9NX13, Q9Y511 | Gene names | BCAS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 4. | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.011673 (rank : 96) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SYTL4_HUMAN
|
||||||
| NC score | 0.010921 (rank : 97) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.010418 (rank : 98) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.009800 (rank : 99) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.009645 (rank : 100) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
SL9A1_HUMAN
|
||||||
| NC score | 0.008144 (rank : 101) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19634 | Gene names | SLC9A1, APNH1, NHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 1 (Na(+)/H(+) exchanger 1) (NHE-1) (Solute carrier family 9 member 1) (Na(+)/H(+) antiporter, amiloride- sensitive) (APNH). | |||||
|
ABCAD_HUMAN
|
||||||
| NC score | 0.007887 (rank : 102) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
IGB1B_MOUSE
|
||||||
| NC score | 0.007783 (rank : 103) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ29 | Gene names | Igbp1b | |||
|
Domain Architecture |

|
|||||
| Description | Immunoglobulin-binding protein 1b (CD79a-binding protein 1b) (Alpha 4- b protein) (Alpha4-b). | |||||
|
GTR2_HUMAN
|
||||||
| NC score | 0.006517 (rank : 104) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11168 | Gene names | SLC2A2, GLUT2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 2 (Glucose transporter type 2, liver). | |||||
|
RBMS1_HUMAN
|
||||||
| NC score | 0.005321 (rank : 105) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
AOF1_MOUSE
|
||||||
| NC score | 0.005236 (rank : 106) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.005115 (rank : 107) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.005064 (rank : 108) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
PANK2_HUMAN
|
||||||
| NC score | 0.004965 (rank : 109) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
KGP2_HUMAN
|
||||||
| NC score | 0.002976 (rank : 110) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.001576 (rank : 111) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
HIPK1_HUMAN
|
||||||
| NC score | 0.000921 (rank : 112) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86Z02, O75125, Q8IYD7, Q8NDN5, Q8NEB6, Q8TBZ1 | Gene names | HIPK1, KIAA0630 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1). | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.000760 (rank : 113) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
KKCC1_MOUSE
|
||||||
| NC score | 0.000454 (rank : 114) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VBY2, Q9R054 | Gene names | Camkk1, Camkk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||