Please be patient as the page loads
|
RRP44_HUMAN
|
||||||
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RRP44_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
PR285_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 2) | NC score | 0.362803 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
REST_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 3) | NC score | 0.048226 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
REST_HUMAN
|
||||||
| θ value | 0.279714 (rank : 4) | NC score | 0.040793 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 0.62314 (rank : 5) | NC score | 0.045077 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.056188 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.055481 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
IPO11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.066357 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UI26, Q8N5R2, Q9NSJ6, Q9NVB1 | Gene names | IPO11, RANBP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-11 (Imp11) (Ran-binding protein 11) (RanBP11). | |||||
|
IPO11_MOUSE
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.064847 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2V6, Q8BU45, Q8K0B4 | Gene names | Ipo11, Ranbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-11 (Imp11) (Ran-binding protein 11) (RanBP11). | |||||
|
G6PD2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.033948 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97324 | Gene names | G6pd2, G6pd-2 | |||
|
Domain Architecture |

|
|||||
| Description | Glucose-6-phosphate 1-dehydrogenase 2 (EC 1.1.1.49) (G6PD). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.055342 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
ZN33A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.003857 (rank : 44) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06730, Q5VZ86 | Gene names | ZNF33A, KIAA0065, KOX31, ZNF33 | |||
|
Domain Architecture |
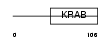
|
|||||
| Description | Zinc finger protein 33A (Zinc finger protein KOX31) (Zinc finger and ZAK-associated protein with KRAB domain) (ZZaPK). | |||||
|
ITPR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.027200 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.055012 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.055311 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
TAIP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.036086 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYN3 | Gene names | TAIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
ZN11B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.003591 (rank : 45) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.033936 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
ITPR3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.026450 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
EXO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.025017 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.021508 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.055773 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.022071 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
ERN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.004135 (rank : 43) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76MJ5, Q6ZNC0 | Gene names | ERN2, IRE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase/endoribonuclease IRE2 precursor (Inositol-requiring protein 2) (hIRE2p) (IRE1b) (Ire1-beta) (Endoplasmic reticulum-to-nucleus signaling 2) [Includes: Serine/threonine-protein kinase (EC 2.7.11.1); Endoribonuclease (EC 3.1.26.-)]. | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.012730 (rank : 40) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
ROBO4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.009374 (rank : 42) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |
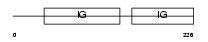
|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
SIN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.033449 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BPZ7, Q00426 | Gene names | MAPKAP1, SIN1 | |||
|
Domain Architecture |
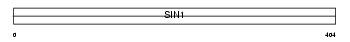
|
|||||
| Description | Stress-activated map kinase-interacting protein 1 (SAPK-interacting protein 1) (Putative Ras inhibitor JC310). | |||||
|
TAF1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.024914 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15573, Q9NWA1 | Gene names | TAF1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit A (TATA box-binding protein-associated factor 1A) (TBP-associated factor RNA polymerase I 48 kDa) (TAFI48) (Transcription factor SL1). | |||||
|
GRPE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.019836 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAA5 | Gene names | GRPEL2 | |||
|
Domain Architecture |
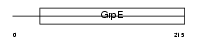
|
|||||
| Description | GrpE protein homolog 2, mitochondrial precursor (Mt-GrpE#2). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.010598 (rank : 41) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.014949 (rank : 39) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.016748 (rank : 38) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
TAI12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.029676 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
TAI12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.029612 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.016750 (rank : 37) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
HELZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.114908 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
M10L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.058011 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9BXT6, Q5TGD5, Q8NBD4, Q9NXW3, Q9UFB3, Q9UGX9 | Gene names | MOV10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1). | |||||
|
M10L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.057553 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q99MV5, Q7TPA9, Q8C3W0, Q924C2 | Gene names | Mov10l1, Champ | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1) (Cardiac helicase activated by MEF2 protein) (Cardiac-specific RNA helicase). | |||||
|
MOV10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.053788 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9HCE1, Q8TEF0, Q9BSY3, Q9BUJ9 | Gene names | MOV10, KIAA1631 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
MOV10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.054228 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P23249, Q9DC64 | Gene names | Mov10, Gb110 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
RENT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.067740 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
RENT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.067662 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
SETX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.056015 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.057291 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.061791 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
RRP44_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
PR285_HUMAN
|
||||||
| NC score | 0.362803 (rank : 2) | θ value | 3.07116e-24 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.114908 (rank : 3) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
RENT1_HUMAN
|
||||||
| NC score | 0.067740 (rank : 4) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
RENT1_MOUSE
|
||||||
| NC score | 0.067662 (rank : 5) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
IPO11_HUMAN
|
||||||
| NC score | 0.066357 (rank : 6) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UI26, Q8N5R2, Q9NSJ6, Q9NVB1 | Gene names | IPO11, RANBP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-11 (Imp11) (Ran-binding protein 11) (RanBP11). | |||||
|
IPO11_MOUSE
|
||||||
| NC score | 0.064847 (rank : 7) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2V6, Q8BU45, Q8K0B4 | Gene names | Ipo11, Ranbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-11 (Imp11) (Ran-binding protein 11) (RanBP11). | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.061791 (rank : 8) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
M10L1_HUMAN
|
||||||
| NC score | 0.058011 (rank : 9) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9BXT6, Q5TGD5, Q8NBD4, Q9NXW3, Q9UFB3, Q9UGX9 | Gene names | MOV10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1). | |||||
|
M10L1_MOUSE
|
||||||
| NC score | 0.057553 (rank : 10) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q99MV5, Q7TPA9, Q8C3W0, Q924C2 | Gene names | Mov10l1, Champ | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1) (Cardiac helicase activated by MEF2 protein) (Cardiac-specific RNA helicase). | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.057291 (rank : 11) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.056188 (rank : 12) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.056015 (rank : 13) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.055773 (rank : 14) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.055481 (rank : 15) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.055342 (rank : 16) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.055311 (rank : 17) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.055012 (rank : 18) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
MOV10_MOUSE
|
||||||
| NC score | 0.054228 (rank : 19) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P23249, Q9DC64 | Gene names | Mov10, Gb110 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
MOV10_HUMAN
|
||||||
| NC score | 0.053788 (rank : 20) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9HCE1, Q8TEF0, Q9BSY3, Q9BUJ9 | Gene names | MOV10, KIAA1631 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.048226 (rank : 21) | θ value | 0.00665767 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.045077 (rank : 22) | θ value | 0.62314 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.040793 (rank : 23) | θ value | 0.279714 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
TAIP2_HUMAN
|
||||||
| NC score | 0.036086 (rank : 24) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYN3 | Gene names | TAIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
G6PD2_MOUSE
|
||||||
| NC score | 0.033948 (rank : 25) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97324 | Gene names | G6pd2, G6pd-2 | |||
|
Domain Architecture |

|
|||||
| Description | Glucose-6-phosphate 1-dehydrogenase 2 (EC 1.1.1.49) (G6PD). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.033936 (rank : 26) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SIN1_HUMAN
|
||||||
| NC score | 0.033449 (rank : 27) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BPZ7, Q00426 | Gene names | MAPKAP1, SIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-activated map kinase-interacting protein 1 (SAPK-interacting protein 1) (Putative Ras inhibitor JC310). | |||||
|
TAI12_HUMAN
|
||||||
| NC score | 0.029676 (rank : 28) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
TAI12_MOUSE
|
||||||
| NC score | 0.029612 (rank : 29) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
ITPR3_HUMAN
|
||||||
| NC score | 0.027200 (rank : 30) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR3_MOUSE
|
||||||
| NC score | 0.026450 (rank : 31) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
EXO1_HUMAN
|
||||||
| NC score | 0.025017 (rank : 32) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
TAF1A_HUMAN
|
||||||
| NC score | 0.024914 (rank : 33) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15573, Q9NWA1 | Gene names | TAF1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit A (TATA box-binding protein-associated factor 1A) (TBP-associated factor RNA polymerase I 48 kDa) (TAFI48) (Transcription factor SL1). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.022071 (rank : 34) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.021508 (rank : 35) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
GRPE2_HUMAN
|
||||||
| NC score | 0.019836 (rank : 36) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAA5 | Gene names | GRPEL2 | |||
|
Domain Architecture |

|
|||||
| Description | GrpE protein homolog 2, mitochondrial precursor (Mt-GrpE#2). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.016750 (rank : 37) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.016748 (rank : 38) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.014949 (rank : 39) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.012730 (rank : 40) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.010598 (rank : 41) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
ROBO4_MOUSE
|
||||||
| NC score | 0.009374 (rank : 42) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
ERN2_HUMAN
|
||||||
| NC score | 0.004135 (rank : 43) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76MJ5, Q6ZNC0 | Gene names | ERN2, IRE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase/endoribonuclease IRE2 precursor (Inositol-requiring protein 2) (hIRE2p) (IRE1b) (Ire1-beta) (Endoplasmic reticulum-to-nucleus signaling 2) [Includes: Serine/threonine-protein kinase (EC 2.7.11.1); Endoribonuclease (EC 3.1.26.-)]. | |||||
|
ZN33A_HUMAN
|
||||||
| NC score | 0.003857 (rank : 44) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06730, Q5VZ86 | Gene names | ZNF33A, KIAA0065, KOX31, ZNF33 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 33A (Zinc finger protein KOX31) (Zinc finger and ZAK-associated protein with KRAB domain) (ZZaPK). | |||||
|
ZN11B_HUMAN
|
||||||
| NC score | 0.003591 (rank : 45) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||