Please be patient as the page loads
|
EXO1_HUMAN
|
||||||
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EXO1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
EXO1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.955221 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
FEN1_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 3) | NC score | 0.588467 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39748 | Gene names | FEN1 | |||
|
Domain Architecture |
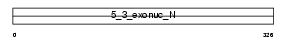
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1) (Maturation factor 1) (MF1) (hFEN-1) (DNase IV). | |||||
|
FEN1_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 4) | NC score | 0.575879 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39749 | Gene names | Fen1, Fen-1 | |||
|
Domain Architecture |
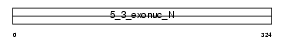
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 5) | NC score | 0.183826 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 6) | NC score | 0.160419 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ERCC5_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 7) | NC score | 0.267943 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 8) | NC score | 0.275983 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 9) | NC score | 0.084314 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
BRCA2_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 10) | NC score | 0.114758 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
EMAL4_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 11) | NC score | 0.060020 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HC35 | Gene names | EML4, C2orf2, EMAPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Echinoderm microtubule-associated protein-like 4 (EMAP-4) (Restrictedly overexpressed proliferation-associated protein) (Ropp 120). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 12) | NC score | 0.169968 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.102450 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.076334 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.093264 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.140845 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
NUDT8_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.078384 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WV74, Q6ZW59 | Gene names | NUDT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 8, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 8). | |||||
|
SYBU_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.053173 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHS8, Q3TQN7, Q401F3, Q401F4, Q571C1, Q6P1J2, Q80XH0, Q8BHS7, Q8BI27 | Gene names | Sybu, Golsyn, Kiaa1472 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntabulin (Syntaxin-1-binding protein) (Golgi-localized syntaphilin- related protein) (m-Golsyn). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.056483 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.086238 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.040360 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
CENPC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.050109 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.017088 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.051827 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.012239 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
RIOK2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.043008 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CQS5, Q91XF3 | Gene names | Riok2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.017576 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
KSR1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.006625 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
RN169_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.041703 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.024912 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.034906 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
CENPR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.044255 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQ82, Q3TLR0, Q3TNR4 | Gene names | Itgb3bp, Cenpr, Nrif3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein R (CENP-R) (Nuclear receptor-interacting factor 3). | |||||
|
M2GD_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.043782 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.011357 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.023482 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
BCL7A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.039421 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.054243 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CS021_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.031148 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D279, Q3UI04 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21 homolog. | |||||
|
M2GD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.041408 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
MAK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.001537 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20794, Q9NUH7 | Gene names | MAK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MAK (EC 2.7.11.22) (Male germ cell- associated kinase). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.050128 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.034107 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
APC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.039098 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.011189 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.015486 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.023586 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.050403 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.025392 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
BCL7A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.034799 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CXE2, Q8C361, Q8C8M8, Q8VD15 | Gene names | Bcl7a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.021707 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CE110_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.022079 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TSH4, Q6A072 | Gene names | Cep110, Cp110, Kiaa0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
DMXL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.014054 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
LIPA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.013500 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75335, O94971 | Gene names | PPFIA4, KIAA0897 | |||
|
Domain Architecture |
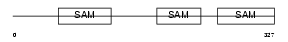
|
|||||
| Description | Liprin-alpha-4 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-4) (PTPRF-interacting protein alpha-4). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.018221 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.033847 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
ABRA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.022549 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.020916 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
DCR1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.018322 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PJP8, Q14701, Q6P5Y3, Q6PKL4 | Gene names | DCLRE1A, KIAA0086, SNM1, SNM1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein (hSNM1) (hSNM1A). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.001368 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.011008 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
IRK14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.010145 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNX9 | Gene names | KCNJ14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-sensitive inward rectifier potassium channel 14 (Potassium channel, inwardly rectifying subfamily J member 14) (Inward rectifier K(+) channel Kir2.4) (IRK4). | |||||
|
MRP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.005780 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15439, Q9Y6J2 | Gene names | ABCC4, MRP4 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance-associated protein 4 (ATP-binding cassette sub- family C member 4) (MRP/cMOAT-related ABC transporter) (Multi-specific organic anion transporter-B) (MOAT-B). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.030172 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RRP44_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.025017 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
RSRC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.029661 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96IZ7, Q96QK2, Q9NZE5 | Gene names | RSRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.054358 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.007432 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
PALB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.016435 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.052339 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TSH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.013701 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.021023 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.034027 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.016546 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
CA026_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.013747 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
CENPU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.024807 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.005875 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
FACD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.014255 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXW9, Q2LA86, Q69YP9, Q6PJN7, Q9BQ06, Q9H9T9 | Gene names | FANCD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group D2 protein (Protein FACD2). | |||||
|
IP3KB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.012178 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
IRAK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.001532 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.018731 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SDK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.001718 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UH53, Q3UFD1, Q6PAL2, Q6V3A4, Q8BMC2, Q8BZI1 | Gene names | Sdk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.023634 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.051922 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
EXO1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
EXO1_MOUSE
|
||||||
| NC score | 0.955221 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
FEN1_HUMAN
|
||||||
| NC score | 0.588467 (rank : 3) | θ value | 8.65492e-19 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39748 | Gene names | FEN1 | |||
|
Domain Architecture |
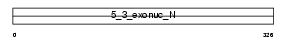
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1) (Maturation factor 1) (MF1) (hFEN-1) (DNase IV). | |||||
|
FEN1_MOUSE
|
||||||
| NC score | 0.575879 (rank : 4) | θ value | 2.13179e-17 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39749 | Gene names | Fen1, Fen-1 | |||
|
Domain Architecture |
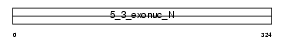
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.275983 (rank : 5) | θ value | 0.000270298 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.267943 (rank : 6) | θ value | 0.000270298 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.183826 (rank : 7) | θ value | 8.40245e-06 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.169968 (rank : 8) | θ value | 0.00665767 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.160419 (rank : 9) | θ value | 3.19293e-05 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.140845 (rank : 10) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
BRCA2_MOUSE
|
||||||
| NC score | 0.114758 (rank : 11) | θ value | 0.00102713 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.102450 (rank : 12) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.093264 (rank : 13) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.086238 (rank : 14) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.084314 (rank : 15) | θ value | 0.00102713 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
NUDT8_HUMAN
|
||||||
| NC score | 0.078384 (rank : 16) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WV74, Q6ZW59 | Gene names | NUDT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 8, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 8). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.076334 (rank : 17) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
EMAL4_HUMAN
|
||||||
| NC score | 0.060020 (rank : 18) | θ value | 0.00298849 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HC35 | Gene names | EML4, C2orf2, EMAPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Echinoderm microtubule-associated protein-like 4 (EMAP-4) (Restrictedly overexpressed proliferation-associated protein) (Ropp 120). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.056483 (rank : 19) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.054358 (rank : 20) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.054243 (rank : 21) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
SYBU_MOUSE
|
||||||
| NC score | 0.053173 (rank : 22) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHS8, Q3TQN7, Q401F3, Q401F4, Q571C1, Q6P1J2, Q80XH0, Q8BHS7, Q8BI27 | Gene names | Sybu, Golsyn, Kiaa1472 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntabulin (Syntaxin-1-binding protein) (Golgi-localized syntaphilin- related protein) (m-Golsyn). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.052339 (rank : 23) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.051922 (rank : 24) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.051827 (rank : 25) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.050403 (rank : 26) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.050128 (rank : 27) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
CENPC_HUMAN
|
||||||
| NC score | 0.050109 (rank : 28) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
CENPR_MOUSE
|
||||||
| NC score | 0.044255 (rank : 29) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQ82, Q3TLR0, Q3TNR4 | Gene names | Itgb3bp, Cenpr, Nrif3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein R (CENP-R) (Nuclear receptor-interacting factor 3). | |||||
|
M2GD_MOUSE
|
||||||
| NC score | 0.043782 (rank : 30) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
RIOK2_MOUSE
|
||||||
| NC score | 0.043008 (rank : 31) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CQS5, Q91XF3 | Gene names | Riok2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
RN169_HUMAN
|
||||||
| NC score | 0.041703 (rank : 32) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
M2GD_HUMAN
|
||||||
| NC score | 0.041408 (rank : 33) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.040360 (rank : 34) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
BCL7A_HUMAN
|
||||||
| NC score | 0.039421 (rank : 35) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.039098 (rank : 36) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.034906 (rank : 37) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BCL7A_MOUSE
|
||||||
| NC score | 0.034799 (rank : 38) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CXE2, Q8C361, Q8C8M8, Q8VD15 | Gene names | Bcl7a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.034107 (rank : 39) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.034027 (rank : 40) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.033847 (rank : 41) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
CS021_MOUSE
|
||||||
| NC score | 0.031148 (rank : 42) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D279, Q3UI04 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21 homolog. | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.030172 (rank : 43) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RSRC1_HUMAN
|
||||||
| NC score | 0.029661 (rank : 44) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96IZ7, Q96QK2, Q9NZE5 | Gene names | RSRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.025392 (rank : 45) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
RRP44_HUMAN
|
||||||
| NC score | 0.025017 (rank : 46) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.024912 (rank : 47) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
CENPU_HUMAN
|
||||||
| NC score | 0.024807 (rank : 48) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.023634 (rank : 49) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.023586 (rank : 50) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.023482 (rank : 51) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
ABRA_HUMAN
|
||||||
| NC score | 0.022549 (rank : 52) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
CE110_MOUSE
|
||||||
| NC score | 0.022079 (rank : 53) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TSH4, Q6A072 | Gene names | Cep110, Cp110, Kiaa0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.021707 (rank : 54) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.021023 (rank : 55) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.020916 (rank : 56) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.018731 (rank : 57) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
DCR1A_HUMAN
|
||||||
| NC score | 0.018322 (rank : 58) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PJP8, Q14701, Q6P5Y3, Q6PKL4 | Gene names | DCLRE1A, KIAA0086, SNM1, SNM1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein (hSNM1) (hSNM1A). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.018221 (rank : 59) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.017576 (rank : 60) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.017088 (rank : 61) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.016546 (rank : 62) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
PALB2_MOUSE
|
||||||
| NC score | 0.016435 (rank : 63) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.015486 (rank : 64) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
FACD2_HUMAN
|
||||||
| NC score | 0.014255 (rank : 65) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXW9, Q2LA86, Q69YP9, Q6PJN7, Q9BQ06, Q9H9T9 | Gene names | FANCD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group D2 protein (Protein FACD2). | |||||
|
DMXL1_HUMAN
|
||||||
| NC score | 0.014054 (rank : 66) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
CA026_HUMAN
|
||||||
| NC score | 0.013747 (rank : 67) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
TSH2_HUMAN
|
||||||
| NC score | 0.013701 (rank : 68) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
LIPA4_HUMAN
|
||||||
| NC score | 0.013500 (rank : 69) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75335, O94971 | Gene names | PPFIA4, KIAA0897 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-4 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-4) (PTPRF-interacting protein alpha-4). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.012239 (rank : 70) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
IP3KB_HUMAN
|
||||||
| NC score | 0.012178 (rank : 71) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.011357 (rank : 72) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.011189 (rank : 73) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.011008 (rank : 74) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
IRK14_HUMAN
|
||||||
| NC score | 0.010145 (rank : 75) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNX9 | Gene names | KCNJ14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-sensitive inward rectifier potassium channel 14 (Potassium channel, inwardly rectifying subfamily J member 14) (Inward rectifier K(+) channel Kir2.4) (IRK4). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.007432 (rank : 76) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
KSR1_MOUSE
|
||||||
| NC score | 0.006625 (rank : 77) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.005875 (rank : 78) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
MRP4_HUMAN
|
||||||
| NC score | 0.005780 (rank : 79) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15439, Q9Y6J2 | Gene names | ABCC4, MRP4 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance-associated protein 4 (ATP-binding cassette sub- family C member 4) (MRP/cMOAT-related ABC transporter) (Multi-specific organic anion transporter-B) (MOAT-B). | |||||
|
SDK1_MOUSE
|
||||||
| NC score | 0.001718 (rank : 80) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UH53, Q3UFD1, Q6PAL2, Q6V3A4, Q8BMC2, Q8BZI1 | Gene names | Sdk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
MAK_HUMAN
|
||||||
| NC score | 0.001537 (rank : 81) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20794, Q9NUH7 | Gene names | MAK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MAK (EC 2.7.11.22) (Male germ cell- associated kinase). | |||||
|
IRAK3_MOUSE
|
||||||
| NC score | 0.001532 (rank : 82) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.001368 (rank : 83) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||