Please be patient as the page loads
|
CA026_HUMAN
|
||||||
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CA026_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
CA026_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.951033 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
EST1A_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 3) | NC score | 0.312018 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 4) | NC score | 0.310077 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 5) | NC score | 0.068035 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
DESP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 6) | NC score | 0.059724 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.072431 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.071501 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.051974 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
RN168_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.065296 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80XJ2 | Gene names | Rnf168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.042040 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.060793 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
NBN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.065895 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.073081 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.049477 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
CB010_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.102043 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z570, Q6ZN26 | Gene names | C2orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf10. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.108197 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
TUFT1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.045447 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.055456 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
DISC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.050442 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRI5, O75045, Q5VT44, Q5VT45, Q8IXJ0, Q8IXJ1, Q9BX19, Q9NRI3, Q9NRI4 | Gene names | DISC1, KIAA0457 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disrupted in schizophrenia 1 protein. | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.054199 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RBBP8_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.061538 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
CUED1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.048185 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWM3, Q9NWD0 | Gene names | CUEDC1 | |||
|
Domain Architecture |
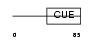
|
|||||
| Description | CUE domain-containing protein 1. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.037497 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.063970 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.051061 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.048548 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.058315 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.036331 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.027341 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.038593 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
SNCAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.027800 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.047434 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.048449 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CCNT1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.025221 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.019097 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
DZIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.047260 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
TR150_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.041651 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
H11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.023653 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |
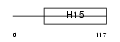
|
|||||
| Description | Histone H1.1. | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.055879 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.050148 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RNF19_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.020211 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NV58, Q9H5H9, Q9H8M8, Q9UFG0, Q9UFX6, Q9Y4Y1 | Gene names | RNF19 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (Dorfin) (Double ring-finger protein) (p38 protein). | |||||
|
SH3G3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.015981 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62421 | Gene names | Sh3gl3, Sh3d2c, Sh3d2c2 | |||
|
Domain Architecture |
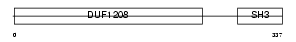
|
|||||
| Description | SH3-containing GRB2-like protein 3 (Endophilin-3) (Endophilin-A3) (SH3 domain protein 2C) (SH3p13). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.068987 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.069229 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.037924 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MCM3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.024321 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.020331 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
RPGR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.025179 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.048091 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.026001 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.028480 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.038513 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.022648 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
NCAM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.007018 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15394 | Gene names | NCAM2, NCAM21 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 2 precursor (N-CAM 2). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.036807 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.017949 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.024201 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
ABCAD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.008528 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.004866 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.034834 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
DEN2C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.011112 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68D51, Q5TCX6, Q6P3R3 | Gene names | DENND2C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.040924 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.040179 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.047645 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.016404 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.012342 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
ARHGG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.008797 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.005649 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.042466 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.037711 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.037566 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.034739 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
PDCD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.018918 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
PSMD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.025761 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43242, Q96EI2, Q9BQA4 | Gene names | PSMD3 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 3 (26S proteasome regulatory subunit S3) (Proteasome subunit p58). | |||||
|
PSMD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.025774 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14685, Q99LL7 | Gene names | Psmd3, P91a, Tstap91a | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 3 (26S proteasome regulatory subunit S3) (Proteasome subunit p58) (Transplantation antigen P91A) (Tum-P91A antigen). | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.031328 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
CDON_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.006631 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4KMG0, O14631 | Gene names | CDON, CDO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
CDON_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.006851 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q32MD9, O88971 | Gene names | Cdon, Cdo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
CJ006_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.019042 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.038849 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
EXO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.013747 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
FA61A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.012367 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8ND56, Q76LX7, Q96AR3, Q96K73, Q96SN5, Q9UFR3 | Gene names | FAM61A, C19orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A (Putative alpha synuclein-binding protein) (AlphaSNBP). | |||||
|
FAS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.010318 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.036557 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SMG5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.051865 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.040774 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ATRX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050692 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
SYCP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.073629 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
CA026_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
CA026_MOUSE
|
||||||
| NC score | 0.951033 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
EST1A_HUMAN
|
||||||
| NC score | 0.312018 (rank : 3) | θ value | 2.36244e-08 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.310077 (rank : 4) | θ value | 6.87365e-08 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.108197 (rank : 5) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CB010_HUMAN
|
||||||
| NC score | 0.102043 (rank : 6) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z570, Q6ZN26 | Gene names | C2orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf10. | |||||
|
SYCP2_HUMAN
|
||||||
| NC score | 0.073629 (rank : 7) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.073081 (rank : 8) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.072431 (rank : 9) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.071501 (rank : 10) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.069229 (rank : 11) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.068987 (rank : 12) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.068035 (rank : 13) | θ value | 0.00035302 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
NBN_HUMAN
|
||||||
| NC score | 0.065895 (rank : 14) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
RN168_MOUSE
|
||||||
| NC score | 0.065296 (rank : 15) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80XJ2 | Gene names | Rnf168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.063970 (rank : 16) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RBBP8_HUMAN
|
||||||
| NC score | 0.061538 (rank : 17) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.060793 (rank : 18) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.059724 (rank : 19) | θ value | 0.00298849 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.058315 (rank : 20) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.055879 (rank : 21) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.055456 (rank : 22) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.054199 (rank : 23) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.051974 (rank : 24) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
SMG5_HUMAN
|
||||||
| NC score | 0.051865 (rank : 25) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.051061 (rank : 26) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.050692 (rank : 27) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
DISC1_HUMAN
|
||||||
| NC score | 0.050442 (rank : 28) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRI5, O75045, Q5VT44, Q5VT45, Q8IXJ0, Q8IXJ1, Q9BX19, Q9NRI3, Q9NRI4 | Gene names | DISC1, KIAA0457 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disrupted in schizophrenia 1 protein. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.050148 (rank : 29) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.049477 (rank : 30) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.048548 (rank : 31) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.048449 (rank : 32) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CUED1_HUMAN
|
||||||
| NC score | 0.048185 (rank : 33) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWM3, Q9NWD0 | Gene names | CUEDC1 | |||
|
Domain Architecture |

|
|||||
| Description | CUE domain-containing protein 1. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.048091 (rank : 34) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.047645 (rank : 35) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.047434 (rank : 36) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
DZIP1_HUMAN
|
||||||
| NC score | 0.047260 (rank : 37) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
TUFT1_HUMAN
|
||||||
| NC score | 0.045447 (rank : 38) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.042466 (rank : 39) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.042040 (rank : 40) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
TR150_HUMAN
|
||||||
| NC score | 0.041651 (rank : 41) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.040924 (rank : 42) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.040774 (rank : 43) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.040179 (rank : 44) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.038849 (rank : 45) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.038593 (rank : 46) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.038513 (rank : 47) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.037924 (rank : 48) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.037711 (rank : 49) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.037566 (rank : 50) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.037497 (rank : 51) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.036807 (rank : 52) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.036557 (rank : 53) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.036331 (rank : 54) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.034834 (rank : 55) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.034739 (rank : 56) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.031328 (rank : 57) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.028480 (rank : 58) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SNCAP_HUMAN
|
||||||
| NC score | 0.027800 (rank : 59) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.027341 (rank : 60) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.026001 (rank : 61) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
PSMD3_MOUSE
|
||||||
| NC score | 0.025774 (rank : 62) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14685, Q99LL7 | Gene names | Psmd3, P91a, Tstap91a | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 3 (26S proteasome regulatory subunit S3) (Proteasome subunit p58) (Transplantation antigen P91A) (Tum-P91A antigen). | |||||
|
PSMD3_HUMAN
|
||||||
| NC score | 0.025761 (rank : 63) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43242, Q96EI2, Q9BQA4 | Gene names | PSMD3 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 3 (26S proteasome regulatory subunit S3) (Proteasome subunit p58). | |||||
|
CCNT1_MOUSE
|
||||||
| NC score | 0.025221 (rank : 64) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
RPGR_HUMAN
|
||||||
| NC score | 0.025179 (rank : 65) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
MCM3A_MOUSE
|
||||||
| NC score | 0.024321 (rank : 66) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.024201 (rank : 67) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
H11_HUMAN
|
||||||
| NC score | 0.023653 (rank : 68) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1. | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.022648 (rank : 69) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.020331 (rank : 70) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
RNF19_HUMAN
|
||||||
| NC score | 0.020211 (rank : 71) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NV58, Q9H5H9, Q9H8M8, Q9UFG0, Q9UFX6, Q9Y4Y1 | Gene names | RNF19 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (Dorfin) (Double ring-finger protein) (p38 protein). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.019097 (rank : 72) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CJ006_MOUSE
|
||||||
| NC score | 0.019042 (rank : 73) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
PDCD2_HUMAN
|
||||||
| NC score | 0.018918 (rank : 74) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.017949 (rank : 75) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.016404 (rank : 76) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SH3G3_MOUSE
|
||||||
| NC score | 0.015981 (rank : 77) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62421 | Gene names | Sh3gl3, Sh3d2c, Sh3d2c2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 3 (Endophilin-3) (Endophilin-A3) (SH3 domain protein 2C) (SH3p13). | |||||
|
EXO1_HUMAN
|
||||||
| NC score | 0.013747 (rank : 78) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
FA61A_HUMAN
|
||||||
| NC score | 0.012367 (rank : 79) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8ND56, Q76LX7, Q96AR3, Q96K73, Q96SN5, Q9UFR3 | Gene names | FAM61A, C19orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A (Putative alpha synuclein-binding protein) (AlphaSNBP). | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.012342 (rank : 80) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
DEN2C_HUMAN
|
||||||
| NC score | 0.011112 (rank : 81) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68D51, Q5TCX6, Q6P3R3 | Gene names | DENND2C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.010318 (rank : 82) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
ARHGG_HUMAN
|
||||||
| NC score | 0.008797 (rank : 83) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
ABCAD_HUMAN
|
||||||
| NC score | 0.008528 (rank : 84) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
NCAM2_HUMAN
|
||||||
| NC score | 0.007018 (rank : 85) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15394 | Gene names | NCAM2, NCAM21 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 2 precursor (N-CAM 2). | |||||
|
CDON_MOUSE
|
||||||
| NC score | 0.006851 (rank : 86) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q32MD9, O88971 | Gene names | Cdon, Cdo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
CDON_HUMAN
|
||||||
| NC score | 0.006631 (rank : 87) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4KMG0, O14631 | Gene names | CDON, CDO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.005649 (rank : 88) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.004866 (rank : 89) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||