Please be patient as the page loads
|
FAS_MOUSE
|
||||||
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FAS_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989188 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
FAS_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
QORX_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 3) | NC score | 0.663596 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q53FA7, O14679, O14685, Q38G78, Q6JLE7, Q9BWB8 | Gene names | TP53I3, PIG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative quinone oxidoreductase (EC 1.-.-.-) (Tumor protein p53- inducible protein 3) (p53-induced protein 3). | |||||
|
OXSM_MOUSE
|
||||||
| θ value | 1.99067e-23 (rank : 4) | NC score | 0.530493 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D404 | Gene names | Oxsm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
OXSM_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 5) | NC score | 0.523513 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NWU1 | Gene names | OXSM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
QOR_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 6) | NC score | 0.573239 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q08257, Q53FT0, Q59EU7, Q6NSK9 | Gene names | CRYZ | |||
|
Domain Architecture |
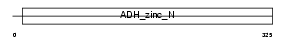
|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
QOR_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 7) | NC score | 0.578890 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P47199, Q62508, Q99L63 | Gene names | Cryz | |||
|
Domain Architecture |
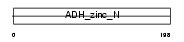
|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
VAT1_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 8) | NC score | 0.535918 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99536, Q13035, Q96A39, Q9BUT8 | Gene names | VAT1 | |||
|
Domain Architecture |
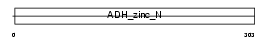
|
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
VAT1_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 9) | NC score | 0.536570 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62465, Q8R3G0, Q8R3T8 | Gene names | Vat1, Vat-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
K1576_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 10) | NC score | 0.528385 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80TB8, Q6PGM9 | Gene names | Kiaa1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
K1576_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 11) | NC score | 0.529142 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HCJ6, Q8IYW8 | Gene names | KIAA1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
ZADH2_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 12) | NC score | 0.522013 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC4 | Gene names | Zadh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
ZADH2_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 13) | NC score | 0.511460 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8N4Q0 | Gene names | ZADH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
LTB4D_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 14) | NC score | 0.371258 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14914, Q8IYQ0, Q9H1X6 | Gene names | LTB4DH | |||
|
Domain Architecture |
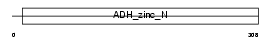
|
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
QORL_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 15) | NC score | 0.409916 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q921W4, Q3UKX2, Q8VCS1, Q9CWR9 | Gene names | Cryzl1 | |||
|
Domain Architecture |
|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog). | |||||
|
MECR_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 16) | NC score | 0.418426 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BV79, Q5SYU0, Q5SYU1, Q5SYU2, Q6IBU9, Q9Y373 | Gene names | MECR, NBRF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38) (HsNrbf-1) (NRBF-1). | |||||
|
QORL_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 17) | NC score | 0.406961 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95825, Q96DY0, Q9NVY7 | Gene names | CRYZL1 | |||
|
Domain Architecture |
|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog) (4P11). | |||||
|
LTB4D_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 18) | NC score | 0.379232 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91YR9, Q3TKT6, Q9CPS1 | Gene names | Ltb4dh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
FABD_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 19) | NC score | 0.196890 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IVS2, O95510, O95511 | Gene names | MCAT, MT | |||
|
Domain Architecture |
|
|||||
| Description | Malonyl CoA-acyl carrier protein transacylase, mitochondrial precursor (EC 2.3.1.39) (MCT) (Mitochondrial malonyltransferase). | |||||
|
MECR_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 20) | NC score | 0.399643 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DCS3, Q99L39 | Gene names | Mecr, Nrbf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38). | |||||
|
DHB4_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 21) | NC score | 0.096451 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
RT4I1_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 22) | NC score | 0.356884 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924D0, Q8R1T0 | Gene names | Rtn4ip1, Nimp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
ADH1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.107875 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00329 | Gene names | Adh1, Adh-1 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1 (EC 1.1.1.1) (Alcohol dehydrogenase A subunit) (ADH-A2). | |||||
|
RT4I1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.327953 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.012464 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
ANR38_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.017631 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9J5, Q8BV03 | Gene names | Ankrd38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
DECR2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.036501 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NUI1, Q6ZRS7, Q96ET0 | Gene names | DECR2, PDCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2) (pDCR). | |||||
|
ZADH1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.145847 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8N7, Q6MZH8 | Gene names | ZADH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
ADH4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.089721 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08319, Q8TCD7 | Gene names | ADH4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II pi chain). | |||||
|
ECHB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.047958 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JY0, Q3TEH9, Q8BJI5, Q8BJM0, Q8BK52 | Gene names | Hadhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trifunctional enzyme subunit beta, mitochondrial precursor (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)]. | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.007876 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.000892 (rank : 84) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
ATS2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.010408 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
DHRS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.035425 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
DHRS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.039788 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
NNMT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.027085 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40261 | Gene names | NNMT | |||
|
Domain Architecture |
|
|||||
| Description | Nicotinamide N-methyltransferase (EC 2.1.1.1). | |||||
|
RDH12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.024063 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
COPB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.011473 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35606 | Gene names | COPB2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta' (Beta'-coat protein) (Beta'-COP) (p102). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.018052 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
APOA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.009038 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
MMTA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.031237 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LX5, Q9CSK6 | Gene names | Mmtag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 homolog. | |||||
|
PPOX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.024522 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
TDRD6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.039594 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
TLR5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.009931 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60602, O15456 | Gene names | TLR5, TIL3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 5 precursor (Toll/interleukin-1 receptor-like protein 3). | |||||
|
ZN553_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | -0.000705 (rank : 87) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3US17, Q3UFP3, Q8R0V0, Q921H7 | Gene names | Znf553, Zfp553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 553. | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.028363 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
BIRC6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.014881 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
DDR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.000172 (rank : 85) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16832 | Gene names | DDR2, NTRKR3, TKT, TYRO10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
DHRS4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.023683 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
RDH12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.019382 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NR8, Q8TAW6 | Gene names | RDH12 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-) (All-trans and 9-cis retinol dehydrogenase). | |||||
|
ATS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.008269 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
COPB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.009238 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55029, Q3U5Z9 | Gene names | Copb2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta' (Beta'-coat protein) (Beta'-COP) (p102). | |||||
|
FANCG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.017456 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15287 | Gene names | FANCG, XRCC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein (Protein FACG) (DNA-repair protein XRCC9). | |||||
|
FIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.034215 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
MTF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | -0.000722 (rank : 88) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
NEP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.008416 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61391, Q6NXX5, Q8K251 | Gene names | Mme | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (CD10 antigen). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.009548 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.009224 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.013820 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.007521 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
ZP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.012749 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
CN135_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.018639 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q63HM2, Q9BQG8, Q9H9F2 | Gene names | C14orf135, FBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf135 precursor (Hepatitis C virus F protein-binding protein 2) (HCV F protein-binding protein 2). | |||||
|
ECHB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.031689 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55084, O14969, Q53TA6, Q96C77, Q9H3F5 | Gene names | HADHB | |||
|
Domain Architecture |
|
|||||
| Description | Trifunctional enzyme subunit beta, mitochondrial precursor (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)]. | |||||
|
INMT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.026119 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95050, Q3KP49, Q9P1Y2, Q9UBY4, Q9UHQ0 | Gene names | INMT | |||
|
Domain Architecture |
|
|||||
| Description | Indolethylamine N-methyltransferase (EC 2.1.1.49) (Aromatic alkylamine N-methyltransferase) (Indolamine N-methyltransferase) (Arylamine N- methyltransferase) (Amine N-methyltransferase). | |||||
|
NU160_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.012502 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12769, Q96GB3, Q9H660 | Gene names | NUP160, KIAA0197, NUP120 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup160 (Nucleoporin Nup160) (160 kDa nucleoporin). | |||||
|
SAST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.044769 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NV23 | Gene names | OLAH, THEDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | S-acyl fatty acid synthase thioesterase, medium chain (EC 3.1.2.14) (Oleoyl-ACP hydrolase) (Thioesterase II) (Thioesterase domain- containing protein 1). | |||||
|
CA026_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.010318 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
GLE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.007434 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
GPR64_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.003417 (rank : 82) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
HCD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.021882 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
ITIH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.006353 (rank : 79) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
K1849_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.005875 (rank : 81) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
KCRS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.006221 (rank : 80) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17540, Q8N1E1 | Gene names | CKMT2 | |||
|
Domain Architecture |

|
|||||
| Description | Creatine kinase, sarcomeric mitochondrial precursor (EC 2.7.3.2) (S- MtCK) (Mib-CK) (Basic-type mitochondrial creatine kinase). | |||||
|
RAB36_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.001258 (rank : 83) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95755, Q7Z4A9, Q9UHP5 | Gene names | RAB36 | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-36. | |||||
|
RTDR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.017045 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
ZBTB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | -0.000334 (rank : 86) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N680, Q5SZ81, Q9P245 | Gene names | ZBTB2, KIAA1483, ZNF437 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 2. | |||||
|
ADH1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.101979 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07327 | Gene names | ADH1A, ADH1 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1A (EC 1.1.1.1) (Alcohol dehydrogenase alpha subunit). | |||||
|
ADH1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.100567 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00325, Q13711, Q4ZGI9, Q96KI7 | Gene names | ADH1B, ADH2 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1B (EC 1.1.1.1) (Alcohol dehydrogenase beta subunit). | |||||
|
ADH1G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.096260 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P00326, Q4PJ18, Q5WRV0, Q6LBW4, Q6NWV0, Q6NZA7 | Gene names | ADH1C, ADH3 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1C (EC 1.1.1.1) (Alcohol dehydrogenase gamma subunit). | |||||
|
ADH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.054176 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYY9 | Gene names | Adh4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II) (Alcohol dehydrogenase II) (ADH2). | |||||
|
ADH6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.067685 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P28332, Q58F53 | Gene names | ADH6 | |||
|
Domain Architecture |
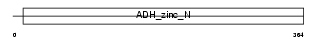
|
|||||
| Description | Alcohol dehydrogenase 6 (EC 1.1.1.1). | |||||
|
ADH7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.070247 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40394, Q13713 | Gene names | ADH7 | |||
|
Domain Architecture |
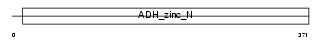
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase). | |||||
|
ADH7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.056199 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64437 | Gene names | Adh7, Adh-3, Adh3 | |||
|
Domain Architecture |
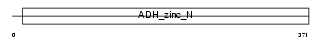
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase) (ADH-C2) (Alcohol dehydrogenase 7). | |||||
|
ADHX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.073102 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11766, Q6FHR2 | Gene names | ADH5, ADHX, FDH | |||
|
Domain Architecture |
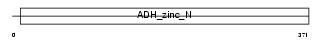
|
|||||
| Description | Alcohol dehydrogenase class 3 chi chain (EC 1.1.1.1) (Alcohol dehydrogenase class III chi chain) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH). | |||||
|
ADHX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.071921 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P28474, Q3TW83, Q8C662 | Gene names | Adh5, Adh-2, Adh2 | |||
|
Domain Architecture |
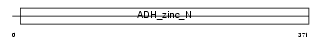
|
|||||
| Description | Alcohol dehydrogenase class 3 (EC 1.1.1.1) (Alcohol dehydrogenase class III) (Alcohol dehydrogenase 2) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH) (FALDH) (Alcohol dehydrogenase-B2) (ADH-B2). | |||||
|
DHSO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.141230 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00796, Q16682, Q9UMD6 | Gene names | SORD | |||
|
Domain Architecture |
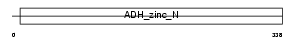
|
|||||
| Description | Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase). | |||||
|
FABD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.092302 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R3F5, Q4FZH0 | Gene names | Mcat, Mt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malonyl CoA-acyl carrier protein transacylase, mitochondrial precursor (EC 2.3.1.39) (MCT) (Mitochondrial malonyltransferase). | |||||
|
ZADH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.115239 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDQ1, Q8BZA2, Q9D1W8 | Gene names | Zadh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
FAS_HUMAN
|
||||||
| NC score | 0.989188 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
QORX_HUMAN
|
||||||
| NC score | 0.663596 (rank : 3) | θ value | 1.37858e-24 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q53FA7, O14679, O14685, Q38G78, Q6JLE7, Q9BWB8 | Gene names | TP53I3, PIG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative quinone oxidoreductase (EC 1.-.-.-) (Tumor protein p53- inducible protein 3) (p53-induced protein 3). | |||||
|
QOR_MOUSE
|
||||||
| NC score | 0.578890 (rank : 4) | θ value | 2.7842e-17 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P47199, Q62508, Q99L63 | Gene names | Cryz | |||
|
Domain Architecture |

|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
QOR_HUMAN
|
||||||
| NC score | 0.573239 (rank : 5) | θ value | 1.13037e-18 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q08257, Q53FT0, Q59EU7, Q6NSK9 | Gene names | CRYZ | |||
|
Domain Architecture |
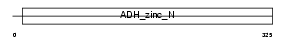
|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
VAT1_MOUSE
|
||||||
| NC score | 0.536570 (rank : 6) | θ value | 3.18553e-13 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62465, Q8R3G0, Q8R3T8 | Gene names | Vat1, Vat-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
VAT1_HUMAN
|
||||||
| NC score | 0.535918 (rank : 7) | θ value | 1.42992e-13 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99536, Q13035, Q96A39, Q9BUT8 | Gene names | VAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
OXSM_MOUSE
|
||||||
| NC score | 0.530493 (rank : 8) | θ value | 1.99067e-23 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D404 | Gene names | Oxsm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
K1576_HUMAN
|
||||||
| NC score | 0.529142 (rank : 9) | θ value | 1.2105e-12 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HCJ6, Q8IYW8 | Gene names | KIAA1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
K1576_MOUSE
|
||||||
| NC score | 0.528385 (rank : 10) | θ value | 9.26847e-13 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80TB8, Q6PGM9 | Gene names | Kiaa1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
OXSM_HUMAN
|
||||||
| NC score | 0.523513 (rank : 11) | θ value | 1.86321e-21 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NWU1 | Gene names | OXSM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
ZADH2_MOUSE
|
||||||
| NC score | 0.522013 (rank : 12) | θ value | 6.00763e-12 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC4 | Gene names | Zadh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
ZADH2_HUMAN
|
||||||
| NC score | 0.511460 (rank : 13) | θ value | 1.02475e-11 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8N4Q0 | Gene names | ZADH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
MECR_HUMAN
|
||||||
| NC score | 0.418426 (rank : 14) | θ value | 7.59969e-07 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BV79, Q5SYU0, Q5SYU1, Q5SYU2, Q6IBU9, Q9Y373 | Gene names | MECR, NBRF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38) (HsNrbf-1) (NRBF-1). | |||||
|
QORL_MOUSE
|
||||||
| NC score | 0.409916 (rank : 15) | θ value | 4.45536e-07 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q921W4, Q3UKX2, Q8VCS1, Q9CWR9 | Gene names | Cryzl1 | |||
|
Domain Architecture |

|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog). | |||||
|
QORL_HUMAN
|
||||||
| NC score | 0.406961 (rank : 16) | θ value | 1.69304e-06 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95825, Q96DY0, Q9NVY7 | Gene names | CRYZL1 | |||
|
Domain Architecture |

|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog) (4P11). | |||||
|
MECR_MOUSE
|
||||||
| NC score | 0.399643 (rank : 17) | θ value | 0.000121331 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DCS3, Q99L39 | Gene names | Mecr, Nrbf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38). | |||||
|
LTB4D_MOUSE
|
||||||
| NC score | 0.379232 (rank : 18) | θ value | 2.88788e-06 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91YR9, Q3TKT6, Q9CPS1 | Gene names | Ltb4dh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
LTB4D_HUMAN
|
||||||
| NC score | 0.371258 (rank : 19) | θ value | 4.45536e-07 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14914, Q8IYQ0, Q9H1X6 | Gene names | LTB4DH | |||
|
Domain Architecture |
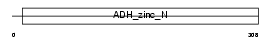
|
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
RT4I1_MOUSE
|
||||||
| NC score | 0.356884 (rank : 20) | θ value | 0.000602161 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924D0, Q8R1T0 | Gene names | Rtn4ip1, Nimp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
RT4I1_HUMAN
|
||||||
| NC score | 0.327953 (rank : 21) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
FABD_HUMAN
|
||||||
| NC score | 0.196890 (rank : 22) | θ value | 1.43324e-05 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IVS2, O95510, O95511 | Gene names | MCAT, MT | |||
|
Domain Architecture |

|
|||||
| Description | Malonyl CoA-acyl carrier protein transacylase, mitochondrial precursor (EC 2.3.1.39) (MCT) (Mitochondrial malonyltransferase). | |||||
|
ZADH1_HUMAN
|
||||||
| NC score | 0.145847 (rank : 23) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8N7, Q6MZH8 | Gene names | ZADH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
DHSO_HUMAN
|
||||||
| NC score | 0.141230 (rank : 24) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00796, Q16682, Q9UMD6 | Gene names | SORD | |||
|
Domain Architecture |
|
|||||
| Description | Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase). | |||||
|
ZADH1_MOUSE
|
||||||
| NC score | 0.115239 (rank : 25) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDQ1, Q8BZA2, Q9D1W8 | Gene names | Zadh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
ADH1_MOUSE
|
||||||
| NC score | 0.107875 (rank : 26) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00329 | Gene names | Adh1, Adh-1 | |||
|
Domain Architecture |

|
|||||
| Description | Alcohol dehydrogenase 1 (EC 1.1.1.1) (Alcohol dehydrogenase A subunit) (ADH-A2). | |||||
|
ADH1A_HUMAN
|
||||||
| NC score | 0.101979 (rank : 27) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07327 | Gene names | ADH1A, ADH1 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1A (EC 1.1.1.1) (Alcohol dehydrogenase alpha subunit). | |||||
|
ADH1B_HUMAN
|
||||||
| NC score | 0.100567 (rank : 28) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00325, Q13711, Q4ZGI9, Q96KI7 | Gene names | ADH1B, ADH2 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1B (EC 1.1.1.1) (Alcohol dehydrogenase beta subunit). | |||||
|
DHB4_MOUSE
|
||||||
| NC score | 0.096451 (rank : 29) | θ value | 0.000158464 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
ADH1G_HUMAN
|
||||||
| NC score | 0.096260 (rank : 30) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P00326, Q4PJ18, Q5WRV0, Q6LBW4, Q6NWV0, Q6NZA7 | Gene names | ADH1C, ADH3 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1C (EC 1.1.1.1) (Alcohol dehydrogenase gamma subunit). | |||||
|
FABD_MOUSE
|
||||||
| NC score | 0.092302 (rank : 31) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R3F5, Q4FZH0 | Gene names | Mcat, Mt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malonyl CoA-acyl carrier protein transacylase, mitochondrial precursor (EC 2.3.1.39) (MCT) (Mitochondrial malonyltransferase). | |||||
|
ADH4_HUMAN
|
||||||
| NC score | 0.089721 (rank : 32) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08319, Q8TCD7 | Gene names | ADH4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II pi chain). | |||||
|
ADHX_HUMAN
|
||||||
| NC score | 0.073102 (rank : 33) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11766, Q6FHR2 | Gene names | ADH5, ADHX, FDH | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 3 chi chain (EC 1.1.1.1) (Alcohol dehydrogenase class III chi chain) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH). | |||||
|
ADHX_MOUSE
|
||||||
| NC score | 0.071921 (rank : 34) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P28474, Q3TW83, Q8C662 | Gene names | Adh5, Adh-2, Adh2 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 3 (EC 1.1.1.1) (Alcohol dehydrogenase class III) (Alcohol dehydrogenase 2) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH) (FALDH) (Alcohol dehydrogenase-B2) (ADH-B2). | |||||
|
ADH7_HUMAN
|
||||||
| NC score | 0.070247 (rank : 35) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40394, Q13713 | Gene names | ADH7 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase). | |||||
|
ADH6_HUMAN
|
||||||
| NC score | 0.067685 (rank : 36) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P28332, Q58F53 | Gene names | ADH6 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 6 (EC 1.1.1.1). | |||||
|
ADH7_MOUSE
|
||||||
| NC score | 0.056199 (rank : 37) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64437 | Gene names | Adh7, Adh-3, Adh3 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase) (ADH-C2) (Alcohol dehydrogenase 7). | |||||
|
ADH4_MOUSE
|
||||||
| NC score | 0.054176 (rank : 38) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYY9 | Gene names | Adh4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II) (Alcohol dehydrogenase II) (ADH2). | |||||
|
ECHB_MOUSE
|
||||||
| NC score | 0.047958 (rank : 39) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JY0, Q3TEH9, Q8BJI5, Q8BJM0, Q8BK52 | Gene names | Hadhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trifunctional enzyme subunit beta, mitochondrial precursor (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)]. | |||||
|
SAST_HUMAN
|
||||||
| NC score | 0.044769 (rank : 40) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NV23 | Gene names | OLAH, THEDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | S-acyl fatty acid synthase thioesterase, medium chain (EC 3.1.2.14) (Oleoyl-ACP hydrolase) (Thioesterase II) (Thioesterase domain- containing protein 1). | |||||
|
DHRS3_MOUSE
|
||||||
| NC score | 0.039788 (rank : 41) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
TDRD6_MOUSE
|
||||||
| NC score | 0.039594 (rank : 42) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
DECR2_HUMAN
|
||||||
| NC score | 0.036501 (rank : 43) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NUI1, Q6ZRS7, Q96ET0 | Gene names | DECR2, PDCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2) (pDCR). | |||||
|
DHRS3_HUMAN
|
||||||
| NC score | 0.035425 (rank : 44) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
FIP1_HUMAN
|
||||||
| NC score | 0.034215 (rank : 45) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
ECHB_HUMAN
|
||||||
| NC score | 0.031689 (rank : 46) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55084, O14969, Q53TA6, Q96C77, Q9H3F5 | Gene names | HADHB | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit beta, mitochondrial precursor (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)]. | |||||
|
MMTA2_MOUSE
|
||||||
| NC score | 0.031237 (rank : 47) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LX5, Q9CSK6 | Gene names | Mmtag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 homolog. | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.028363 (rank : 48) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
NNMT_HUMAN
|
||||||
| NC score | 0.027085 (rank : 49) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40261 | Gene names | NNMT | |||
|
Domain Architecture |

|
|||||
| Description | Nicotinamide N-methyltransferase (EC 2.1.1.1). | |||||
|
INMT_HUMAN
|
||||||
| NC score | 0.026119 (rank : 50) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95050, Q3KP49, Q9P1Y2, Q9UBY4, Q9UHQ0 | Gene names | INMT | |||
|
Domain Architecture |

|
|||||
| Description | Indolethylamine N-methyltransferase (EC 2.1.1.49) (Aromatic alkylamine N-methyltransferase) (Indolamine N-methyltransferase) (Arylamine N- methyltransferase) (Amine N-methyltransferase). | |||||
|
PPOX_HUMAN
|
||||||
| NC score | 0.024522 (rank : 51) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
RDH12_MOUSE
|
||||||
| NC score | 0.024063 (rank : 52) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
DHRS4_HUMAN
|
||||||
| NC score | 0.023683 (rank : 53) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
HCD2_HUMAN
|
||||||
| NC score | 0.021882 (rank : 54) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
RDH12_HUMAN
|
||||||
| NC score | 0.019382 (rank : 55) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NR8, Q8TAW6 | Gene names | RDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-) (All-trans and 9-cis retinol dehydrogenase). | |||||
|
CN135_HUMAN
|
||||||
| NC score | 0.018639 (rank : 56) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q63HM2, Q9BQG8, Q9H9F2 | Gene names | C14orf135, FBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf135 precursor (Hepatitis C virus F protein-binding protein 2) (HCV F protein-binding protein 2). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.018052 (rank : 57) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
ANR38_MOUSE
|
||||||
| NC score | 0.017631 (rank : 58) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9J5, Q8BV03 | Gene names | Ankrd38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
FANCG_HUMAN
|
||||||
| NC score | 0.017456 (rank : 59) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15287 | Gene names | FANCG, XRCC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein (Protein FACG) (DNA-repair protein XRCC9). | |||||
|
RTDR1_HUMAN
|
||||||
| NC score | 0.017045 (rank : 60) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
BIRC6_HUMAN
|
||||||
| NC score | 0.014881 (rank : 61) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.013820 (rank : 62) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
ZP2_HUMAN
|
||||||
| NC score | 0.012749 (rank : 63) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
NU160_HUMAN
|
||||||
| NC score | 0.012502 (rank : 64) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12769, Q96GB3, Q9H660 | Gene names | NUP160, KIAA0197, NUP120 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup160 (Nucleoporin Nup160) (160 kDa nucleoporin). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.012464 (rank : 65) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
COPB2_HUMAN
|
||||||
| NC score | 0.011473 (rank : 66) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35606 | Gene names | COPB2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta' (Beta'-coat protein) (Beta'-COP) (p102). | |||||
|
ATS2_HUMAN
|
||||||
| NC score | 0.010408 (rank : 67) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
CA026_HUMAN
|
||||||
| NC score | 0.010318 (rank : 68) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
TLR5_HUMAN
|
||||||
| NC score | 0.009931 (rank : 69) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60602, O15456 | Gene names | TLR5, TIL3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 5 precursor (Toll/interleukin-1 receptor-like protein 3). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.009548 (rank : 70) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
COPB2_MOUSE
|
||||||
| NC score | 0.009238 (rank : 71) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55029, Q3U5Z9 | Gene names | Copb2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta' (Beta'-coat protein) (Beta'-COP) (p102). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.009224 (rank : 72) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
APOA_HUMAN
|
||||||
| NC score | 0.009038 (rank : 73) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
NEP_MOUSE
|
||||||
| NC score | 0.008416 (rank : 74) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61391, Q6NXX5, Q8K251 | Gene names | Mme | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (CD10 antigen). | |||||
|
ATS2_MOUSE
|
||||||
| NC score | 0.008269 (rank : 75) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.007876 (rank : 76) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.007521 (rank : 77) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
GLE1_HUMAN
|
||||||
| NC score | 0.007434 (rank : 78) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.006353 (rank : 79) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
KCRS_HUMAN
|
||||||
| NC score | 0.006221 (rank : 80) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17540, Q8N1E1 | Gene names | CKMT2 | |||
|
Domain Architecture |

|
|||||
| Description | Creatine kinase, sarcomeric mitochondrial precursor (EC 2.7.3.2) (S- MtCK) (Mib-CK) (Basic-type mitochondrial creatine kinase). | |||||
|
K1849_HUMAN
|
||||||
| NC score | 0.005875 (rank : 81) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
GPR64_MOUSE
|
||||||
| NC score | 0.003417 (rank : 82) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
RAB36_HUMAN
|
||||||
| NC score | 0.001258 (rank : 83) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95755, Q7Z4A9, Q9UHP5 | Gene names | RAB36 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-36. | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.000892 (rank : 84) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
DDR2_HUMAN
|
||||||
| NC score | 0.000172 (rank : 85) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16832 | Gene names | DDR2, NTRKR3, TKT, TYRO10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
ZBTB2_HUMAN
|
||||||
| NC score | -0.000334 (rank : 86) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N680, Q5SZ81, Q9P245 | Gene names | ZBTB2, KIAA1483, ZNF437 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 2. | |||||
|
ZN553_MOUSE
|
||||||
| NC score | -0.000705 (rank : 87) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3US17, Q3UFP3, Q8R0V0, Q921H7 | Gene names | Znf553, Zfp553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 553. | |||||
|
MTF1_HUMAN
|
||||||
| NC score | -0.000722 (rank : 88) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||