Please be patient as the page loads
|
FAS_HUMAN
|
||||||
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FAS_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
FAS_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989188 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
OXSM_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 3) | NC score | 0.538354 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D404 | Gene names | Oxsm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
OXSM_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 4) | NC score | 0.531709 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NWU1 | Gene names | OXSM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
QORX_HUMAN
|
||||||
| θ value | 7.56453e-23 (rank : 5) | NC score | 0.651880 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q53FA7, O14679, O14685, Q38G78, Q6JLE7, Q9BWB8 | Gene names | TP53I3, PIG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative quinone oxidoreductase (EC 1.-.-.-) (Tumor protein p53- inducible protein 3) (p53-induced protein 3). | |||||
|
QOR_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 6) | NC score | 0.572392 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08257, Q53FT0, Q59EU7, Q6NSK9 | Gene names | CRYZ | |||
|
Domain Architecture |
|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
QOR_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 7) | NC score | 0.572604 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47199, Q62508, Q99L63 | Gene names | Cryz | |||
|
Domain Architecture |
|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
ZADH2_MOUSE
|
||||||
| θ value | 2.0648e-12 (rank : 8) | NC score | 0.513211 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGC4 | Gene names | Zadh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
K1576_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 9) | NC score | 0.518480 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCJ6, Q8IYW8 | Gene names | KIAA1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
K1576_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 10) | NC score | 0.517674 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80TB8, Q6PGM9 | Gene names | Kiaa1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
ZADH2_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 11) | NC score | 0.503118 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4Q0 | Gene names | ZADH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
VAT1_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 12) | NC score | 0.522728 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62465, Q8R3G0, Q8R3T8 | Gene names | Vat1, Vat-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
VAT1_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 13) | NC score | 0.523413 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99536, Q13035, Q96A39, Q9BUT8 | Gene names | VAT1 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
QORL_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 14) | NC score | 0.406621 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q921W4, Q3UKX2, Q8VCS1, Q9CWR9 | Gene names | Cryzl1 | |||
|
Domain Architecture |
|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog). | |||||
|
QORL_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 15) | NC score | 0.403912 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95825, Q96DY0, Q9NVY7 | Gene names | CRYZL1 | |||
|
Domain Architecture |
|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog) (4P11). | |||||
|
LTB4D_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 16) | NC score | 0.362614 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91YR9, Q3TKT6, Q9CPS1 | Gene names | Ltb4dh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
MECR_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 17) | NC score | 0.426381 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BV79, Q5SYU0, Q5SYU1, Q5SYU2, Q6IBU9, Q9Y373 | Gene names | MECR, NBRF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38) (HsNrbf-1) (NRBF-1). | |||||
|
LTB4D_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 18) | NC score | 0.352474 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14914, Q8IYQ0, Q9H1X6 | Gene names | LTB4DH | |||
|
Domain Architecture |
|
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
FABD_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 19) | NC score | 0.225827 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVS2, O95510, O95511 | Gene names | MCAT, MT | |||
|
Domain Architecture |
|
|||||
| Description | Malonyl CoA-acyl carrier protein transacylase, mitochondrial precursor (EC 2.3.1.39) (MCT) (Mitochondrial malonyltransferase). | |||||
|
MECR_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 20) | NC score | 0.403912 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DCS3, Q99L39 | Gene names | Mecr, Nrbf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38). | |||||
|
ADH1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 21) | NC score | 0.168000 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00329 | Gene names | Adh1, Adh-1 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1 (EC 1.1.1.1) (Alcohol dehydrogenase A subunit) (ADH-A2). | |||||
|
FABD_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 22) | NC score | 0.125105 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3F5, Q4FZH0 | Gene names | Mcat, Mt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malonyl CoA-acyl carrier protein transacylase, mitochondrial precursor (EC 2.3.1.39) (MCT) (Mitochondrial malonyltransferase). | |||||
|
ADH1A_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 23) | NC score | 0.162426 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07327 | Gene names | ADH1A, ADH1 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1A (EC 1.1.1.1) (Alcohol dehydrogenase alpha subunit). | |||||
|
ADH1G_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 24) | NC score | 0.157297 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P00326, Q4PJ18, Q5WRV0, Q6LBW4, Q6NWV0, Q6NZA7 | Gene names | ADH1C, ADH3 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1C (EC 1.1.1.1) (Alcohol dehydrogenase gamma subunit). | |||||
|
ADH4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 25) | NC score | 0.147815 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08319, Q8TCD7 | Gene names | ADH4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II pi chain). | |||||
|
ADH1B_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.161055 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00325, Q13711, Q4ZGI9, Q96KI7 | Gene names | ADH1B, ADH2 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1B (EC 1.1.1.1) (Alcohol dehydrogenase beta subunit). | |||||
|
DHB4_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 27) | NC score | 0.086835 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
RT4I1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 28) | NC score | 0.329224 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924D0, Q8R1T0 | Gene names | Rtn4ip1, Nimp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
HXK3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.031100 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52790 | Gene names | HK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hexokinase-3 (EC 2.7.1.1) (Hexokinase type III) (HK III). | |||||
|
ADH6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.128093 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P28332, Q58F53 | Gene names | ADH6 | |||
|
Domain Architecture |
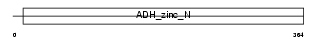
|
|||||
| Description | Alcohol dehydrogenase 6 (EC 1.1.1.1). | |||||
|
ADH7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.131131 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P40394, Q13713 | Gene names | ADH7 | |||
|
Domain Architecture |
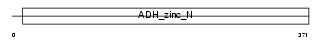
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase). | |||||
|
HXK3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.029626 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TRM8, Q3TAX6, Q3UDP1, Q3UEA4 | Gene names | Hk3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hexokinase-3 (EC 2.7.1.1) (Hexokinase type III) (HK III). | |||||
|
SAST_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.055949 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NV23 | Gene names | OLAH, THEDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | S-acyl fatty acid synthase thioesterase, medium chain (EC 3.1.2.14) (Oleoyl-ACP hydrolase) (Thioesterase II) (Thioesterase domain- containing protein 1). | |||||
|
DHB4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.038945 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
ECHB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.048520 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JY0, Q3TEH9, Q8BJI5, Q8BJM0, Q8BK52 | Gene names | Hadhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trifunctional enzyme subunit beta, mitochondrial precursor (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)]. | |||||
|
APOA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.010307 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
RT4I1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.301466 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
TDRD6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.043746 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
FIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.044380 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
ADH7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.117485 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64437 | Gene names | Adh7, Adh-3, Adh3 | |||
|
Domain Architecture |
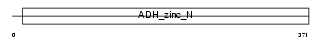
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase) (ADH-C2) (Alcohol dehydrogenase 7). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.002999 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.027745 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
CAN9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.006980 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.007346 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ADHX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.129724 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P28474, Q3TW83, Q8C662 | Gene names | Adh5, Adh-2, Adh2 | |||
|
Domain Architecture |
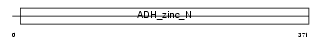
|
|||||
| Description | Alcohol dehydrogenase class 3 (EC 1.1.1.1) (Alcohol dehydrogenase class III) (Alcohol dehydrogenase 2) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH) (FALDH) (Alcohol dehydrogenase-B2) (ADH-B2). | |||||
|
CB032_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.027461 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5M8N0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf32 homolog. | |||||
|
CF165_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.023090 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYR0, Q8N9U4 | Gene names | C6orf165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf165. | |||||
|
FIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.027080 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
SAMH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.016982 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Z3, Q5JXG8, Q9H004, Q9H005, Q9H3U9 | Gene names | SAMHD1, MOP5 | |||
|
Domain Architecture |
|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5). | |||||
|
TLR5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.008801 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60602, O15456 | Gene names | TLR5, TIL3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 5 precursor (Toll/interleukin-1 receptor-like protein 3). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.010123 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.010152 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
CB032_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.025185 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96F85, Q9UFZ0 | Gene names | C2orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf32. | |||||
|
CJ033_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.013605 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
ECHB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.032116 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55084, O14969, Q53TA6, Q96C77, Q9H3F5 | Gene names | HADHB | |||
|
Domain Architecture |
|
|||||
| Description | Trifunctional enzyme subunit beta, mitochondrial precursor (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)]. | |||||
|
INMT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.019294 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95050, Q3KP49, Q9P1Y2, Q9UBY4, Q9UHQ0 | Gene names | INMT | |||
|
Domain Architecture |
|
|||||
| Description | Indolethylamine N-methyltransferase (EC 2.1.1.49) (Aromatic alkylamine N-methyltransferase) (Indolamine N-methyltransferase) (Arylamine N- methyltransferase) (Amine N-methyltransferase). | |||||
|
MIA40_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.019924 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEA4 | Gene names | Chchd4, Mia40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial intermembrane space import and assembly protein 40 (Coiled-coil-helix-coiled-coil-helix domain-containing protein 4). | |||||
|
DHRS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.023895 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
HEM0_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.006989 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22557, Q13735 | Gene names | ALAS2, ALASE, ASB | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, erythroid-specific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta- aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-E). | |||||
|
NAT6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.014144 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q93015, Q93014 | Gene names | NAT6, FUS2 | |||
|
Domain Architecture |
|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
NPM3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.012755 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75607, Q9UNY6 | Gene names | NPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-3. | |||||
|
REC8L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.011853 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95072, Q8WUV8, Q9BTF2, Q9NVQ9 | Gene names | REC8L1, REC8 | |||
|
Domain Architecture |

|
|||||
| Description | Meiotic recombination protein REC8-like 1 (Cohesin Rec8p). | |||||
|
RFPL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.003918 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75678 | Gene names | RFPL2, RNF79 | |||
|
Domain Architecture |
|
|||||
| Description | Ret finger protein-like 2 (RING finger protein 79). | |||||
|
STAT6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.006177 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
SYNJ2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.006098 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.003761 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
ADH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.111514 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYY9 | Gene names | Adh4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II) (Alcohol dehydrogenase II) (ADH2). | |||||
|
ADHX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.130835 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11766, Q6FHR2 | Gene names | ADH5, ADHX, FDH | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 3 chi chain (EC 1.1.1.1) (Alcohol dehydrogenase class III chi chain) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH). | |||||
|
DHSO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.176973 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q00796, Q16682, Q9UMD6 | Gene names | SORD | |||
|
Domain Architecture |
|
|||||
| Description | Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase). | |||||
|
ZADH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.127357 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8N7, Q6MZH8 | Gene names | ZADH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
ZADH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.097932 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDQ1, Q8BZA2, Q9D1W8 | Gene names | Zadh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
FAS_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.989188 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
QORX_HUMAN
|
||||||
| NC score | 0.651880 (rank : 3) | θ value | 7.56453e-23 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q53FA7, O14679, O14685, Q38G78, Q6JLE7, Q9BWB8 | Gene names | TP53I3, PIG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative quinone oxidoreductase (EC 1.-.-.-) (Tumor protein p53- inducible protein 3) (p53-induced protein 3). | |||||
|
QOR_MOUSE
|
||||||
| NC score | 0.572604 (rank : 4) | θ value | 1.99529e-15 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47199, Q62508, Q99L63 | Gene names | Cryz | |||
|
Domain Architecture |

|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
QOR_HUMAN
|
||||||
| NC score | 0.572392 (rank : 5) | θ value | 8.10077e-17 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08257, Q53FT0, Q59EU7, Q6NSK9 | Gene names | CRYZ | |||
|
Domain Architecture |
|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
OXSM_MOUSE
|
||||||
| NC score | 0.538354 (rank : 6) | θ value | 2.59989e-23 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D404 | Gene names | Oxsm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
OXSM_HUMAN
|
||||||
| NC score | 0.531709 (rank : 7) | θ value | 5.79196e-23 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NWU1 | Gene names | OXSM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
VAT1_HUMAN
|
||||||
| NC score | 0.523413 (rank : 8) | θ value | 1.9326e-10 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99536, Q13035, Q96A39, Q9BUT8 | Gene names | VAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
VAT1_MOUSE
|
||||||
| NC score | 0.522728 (rank : 9) | θ value | 5.08577e-11 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62465, Q8R3G0, Q8R3T8 | Gene names | Vat1, Vat-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
K1576_HUMAN
|
||||||
| NC score | 0.518480 (rank : 10) | θ value | 7.84624e-12 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCJ6, Q8IYW8 | Gene names | KIAA1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
K1576_MOUSE
|
||||||
| NC score | 0.517674 (rank : 11) | θ value | 1.02475e-11 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80TB8, Q6PGM9 | Gene names | Kiaa1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
ZADH2_MOUSE
|
||||||
| NC score | 0.513211 (rank : 12) | θ value | 2.0648e-12 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGC4 | Gene names | Zadh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
ZADH2_HUMAN
|
||||||
| NC score | 0.503118 (rank : 13) | θ value | 1.33837e-11 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4Q0 | Gene names | ZADH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
MECR_HUMAN
|
||||||
| NC score | 0.426381 (rank : 14) | θ value | 2.21117e-06 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BV79, Q5SYU0, Q5SYU1, Q5SYU2, Q6IBU9, Q9Y373 | Gene names | MECR, NBRF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38) (HsNrbf-1) (NRBF-1). | |||||
|
QORL_MOUSE
|
||||||
| NC score | 0.406621 (rank : 15) | θ value | 3.41135e-07 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q921W4, Q3UKX2, Q8VCS1, Q9CWR9 | Gene names | Cryzl1 | |||
|
Domain Architecture |

|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog). | |||||
|
MECR_MOUSE
|
||||||
| NC score | 0.403912 (rank : 16) | θ value | 0.000270298 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DCS3, Q99L39 | Gene names | Mecr, Nrbf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38). | |||||
|
QORL_HUMAN
|
||||||
| NC score | 0.403912 (rank : 17) | θ value | 1.29631e-06 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95825, Q96DY0, Q9NVY7 | Gene names | CRYZL1 | |||
|
Domain Architecture |

|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog) (4P11). | |||||
|
LTB4D_MOUSE
|
||||||
| NC score | 0.362614 (rank : 18) | θ value | 2.21117e-06 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91YR9, Q3TKT6, Q9CPS1 | Gene names | Ltb4dh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
LTB4D_HUMAN
|
||||||
| NC score | 0.352474 (rank : 19) | θ value | 1.43324e-05 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14914, Q8IYQ0, Q9H1X6 | Gene names | LTB4DH | |||
|
Domain Architecture |
|
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
RT4I1_MOUSE
|
||||||
| NC score | 0.329224 (rank : 20) | θ value | 0.0431538 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924D0, Q8R1T0 | Gene names | Rtn4ip1, Nimp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
RT4I1_HUMAN
|
||||||
| NC score | 0.301466 (rank : 21) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
FABD_HUMAN
|
||||||
| NC score | 0.225827 (rank : 22) | θ value | 1.87187e-05 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVS2, O95510, O95511 | Gene names | MCAT, MT | |||
|
Domain Architecture |

|
|||||
| Description | Malonyl CoA-acyl carrier protein transacylase, mitochondrial precursor (EC 2.3.1.39) (MCT) (Mitochondrial malonyltransferase). | |||||
|
DHSO_HUMAN
|
||||||
| NC score | 0.176973 (rank : 23) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q00796, Q16682, Q9UMD6 | Gene names | SORD | |||
|
Domain Architecture |
|
|||||
| Description | Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase). | |||||
|
ADH1_MOUSE
|
||||||
| NC score | 0.168000 (rank : 24) | θ value | 0.00665767 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00329 | Gene names | Adh1, Adh-1 | |||
|
Domain Architecture |

|
|||||
| Description | Alcohol dehydrogenase 1 (EC 1.1.1.1) (Alcohol dehydrogenase A subunit) (ADH-A2). | |||||
|
ADH1A_HUMAN
|
||||||
| NC score | 0.162426 (rank : 25) | θ value | 0.0148317 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07327 | Gene names | ADH1A, ADH1 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1A (EC 1.1.1.1) (Alcohol dehydrogenase alpha subunit). | |||||
|
ADH1B_HUMAN
|
||||||
| NC score | 0.161055 (rank : 26) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00325, Q13711, Q4ZGI9, Q96KI7 | Gene names | ADH1B, ADH2 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1B (EC 1.1.1.1) (Alcohol dehydrogenase beta subunit). | |||||
|
ADH1G_HUMAN
|
||||||
| NC score | 0.157297 (rank : 27) | θ value | 0.0193708 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P00326, Q4PJ18, Q5WRV0, Q6LBW4, Q6NWV0, Q6NZA7 | Gene names | ADH1C, ADH3 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1C (EC 1.1.1.1) (Alcohol dehydrogenase gamma subunit). | |||||
|
ADH4_HUMAN
|
||||||
| NC score | 0.147815 (rank : 28) | θ value | 0.0252991 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08319, Q8TCD7 | Gene names | ADH4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II pi chain). | |||||
|
ADH7_HUMAN
|
||||||
| NC score | 0.131131 (rank : 29) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P40394, Q13713 | Gene names | ADH7 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase). | |||||
|
ADHX_HUMAN
|
||||||
| NC score | 0.130835 (rank : 30) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11766, Q6FHR2 | Gene names | ADH5, ADHX, FDH | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 3 chi chain (EC 1.1.1.1) (Alcohol dehydrogenase class III chi chain) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH). | |||||
|
ADHX_MOUSE
|
||||||
| NC score | 0.129724 (rank : 31) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P28474, Q3TW83, Q8C662 | Gene names | Adh5, Adh-2, Adh2 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 3 (EC 1.1.1.1) (Alcohol dehydrogenase class III) (Alcohol dehydrogenase 2) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH) (FALDH) (Alcohol dehydrogenase-B2) (ADH-B2). | |||||
|
ADH6_HUMAN
|
||||||
| NC score | 0.128093 (rank : 32) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P28332, Q58F53 | Gene names | ADH6 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 6 (EC 1.1.1.1). | |||||
|
ZADH1_HUMAN
|
||||||
| NC score | 0.127357 (rank : 33) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8N7, Q6MZH8 | Gene names | ZADH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
FABD_MOUSE
|
||||||
| NC score | 0.125105 (rank : 34) | θ value | 0.00869519 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3F5, Q4FZH0 | Gene names | Mcat, Mt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malonyl CoA-acyl carrier protein transacylase, mitochondrial precursor (EC 2.3.1.39) (MCT) (Mitochondrial malonyltransferase). | |||||
|
ADH7_MOUSE
|
||||||
| NC score | 0.117485 (rank : 35) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64437 | Gene names | Adh7, Adh-3, Adh3 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase) (ADH-C2) (Alcohol dehydrogenase 7). | |||||
|
ADH4_MOUSE
|
||||||
| NC score | 0.111514 (rank : 36) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYY9 | Gene names | Adh4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II) (Alcohol dehydrogenase II) (ADH2). | |||||
|
ZADH1_MOUSE
|
||||||
| NC score | 0.097932 (rank : 37) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDQ1, Q8BZA2, Q9D1W8 | Gene names | Zadh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
DHB4_MOUSE
|
||||||
| NC score | 0.086835 (rank : 38) | θ value | 0.0431538 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
SAST_HUMAN
|
||||||
| NC score | 0.055949 (rank : 39) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NV23 | Gene names | OLAH, THEDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | S-acyl fatty acid synthase thioesterase, medium chain (EC 3.1.2.14) (Oleoyl-ACP hydrolase) (Thioesterase II) (Thioesterase domain- containing protein 1). | |||||
|
ECHB_MOUSE
|
||||||
| NC score | 0.048520 (rank : 40) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JY0, Q3TEH9, Q8BJI5, Q8BJM0, Q8BK52 | Gene names | Hadhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trifunctional enzyme subunit beta, mitochondrial precursor (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)]. | |||||
|
FIP1_HUMAN
|
||||||
| NC score | 0.044380 (rank : 41) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
TDRD6_MOUSE
|
||||||
| NC score | 0.043746 (rank : 42) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
DHB4_HUMAN
|
||||||
| NC score | 0.038945 (rank : 43) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
ECHB_HUMAN
|
||||||
| NC score | 0.032116 (rank : 44) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55084, O14969, Q53TA6, Q96C77, Q9H3F5 | Gene names | HADHB | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit beta, mitochondrial precursor (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)]. | |||||
|
HXK3_HUMAN
|
||||||
| NC score | 0.031100 (rank : 45) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52790 | Gene names | HK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hexokinase-3 (EC 2.7.1.1) (Hexokinase type III) (HK III). | |||||
|
HXK3_MOUSE
|
||||||
| NC score | 0.029626 (rank : 46) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TRM8, Q3TAX6, Q3UDP1, Q3UEA4 | Gene names | Hk3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hexokinase-3 (EC 2.7.1.1) (Hexokinase type III) (HK III). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.027745 (rank : 47) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
CB032_MOUSE
|
||||||
| NC score | 0.027461 (rank : 48) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5M8N0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf32 homolog. | |||||
|
FIP1_MOUSE
|
||||||
| NC score | 0.027080 (rank : 49) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
CB032_HUMAN
|
||||||
| NC score | 0.025185 (rank : 50) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96F85, Q9UFZ0 | Gene names | C2orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf32. | |||||
|
DHRS3_MOUSE
|
||||||
| NC score | 0.023895 (rank : 51) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
CF165_HUMAN
|
||||||
| NC score | 0.023090 (rank : 52) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYR0, Q8N9U4 | Gene names | C6orf165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf165. | |||||
|
MIA40_MOUSE
|
||||||
| NC score | 0.019924 (rank : 53) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEA4 | Gene names | Chchd4, Mia40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial intermembrane space import and assembly protein 40 (Coiled-coil-helix-coiled-coil-helix domain-containing protein 4). | |||||
|
INMT_HUMAN
|
||||||
| NC score | 0.019294 (rank : 54) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95050, Q3KP49, Q9P1Y2, Q9UBY4, Q9UHQ0 | Gene names | INMT | |||
|
Domain Architecture |

|
|||||
| Description | Indolethylamine N-methyltransferase (EC 2.1.1.49) (Aromatic alkylamine N-methyltransferase) (Indolamine N-methyltransferase) (Arylamine N- methyltransferase) (Amine N-methyltransferase). | |||||
|
SAMH1_HUMAN
|
||||||
| NC score | 0.016982 (rank : 55) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Z3, Q5JXG8, Q9H004, Q9H005, Q9H3U9 | Gene names | SAMHD1, MOP5 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5). | |||||
|
NAT6_HUMAN
|
||||||
| NC score | 0.014144 (rank : 56) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q93015, Q93014 | Gene names | NAT6, FUS2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
CJ033_MOUSE
|
||||||
| NC score | 0.013605 (rank : 57) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
NPM3_HUMAN
|
||||||
| NC score | 0.012755 (rank : 58) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75607, Q9UNY6 | Gene names | NPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-3. | |||||
|
REC8L_HUMAN
|
||||||
| NC score | 0.011853 (rank : 59) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95072, Q8WUV8, Q9BTF2, Q9NVQ9 | Gene names | REC8L1, REC8 | |||
|
Domain Architecture |

|
|||||
| Description | Meiotic recombination protein REC8-like 1 (Cohesin Rec8p). | |||||
|
APOA_HUMAN
|
||||||
| NC score | 0.010307 (rank : 60) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.010152 (rank : 61) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.010123 (rank : 62) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
TLR5_HUMAN
|
||||||
| NC score | 0.008801 (rank : 63) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60602, O15456 | Gene names | TLR5, TIL3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 5 precursor (Toll/interleukin-1 receptor-like protein 3). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.007346 (rank : 64) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
HEM0_HUMAN
|
||||||
| NC score | 0.006989 (rank : 65) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22557, Q13735 | Gene names | ALAS2, ALASE, ASB | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, erythroid-specific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta- aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-E). | |||||
|
CAN9_MOUSE
|
||||||
| NC score | 0.006980 (rank : 66) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
STAT6_HUMAN
|
||||||
| NC score | 0.006177 (rank : 67) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
SYNJ2_MOUSE
|
||||||
| NC score | 0.006098 (rank : 68) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
RFPL2_HUMAN
|
||||||
| NC score | 0.003918 (rank : 69) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75678 | Gene names | RFPL2, RNF79 | |||
|
Domain Architecture |

|
|||||
| Description | Ret finger protein-like 2 (RING finger protein 79). | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.003761 (rank : 70) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.002999 (rank : 71) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||