Please be patient as the page loads
|
NAT6_HUMAN
|
||||||
| SwissProt Accessions | Q93015, Q93014 | Gene names | NAT6, FUS2 | |||
|
Domain Architecture |
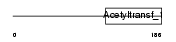
|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NAT6_HUMAN
|
||||||
| θ value | 2.20485e-131 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q93015, Q93014 | Gene names | NAT6, FUS2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
NAT6_MOUSE
|
||||||
| θ value | 2.01746e-92 (rank : 2) | NC score | 0.968418 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R123, Q8QZY5, Q8R014, Q9ERM0 | Gene names | Nat6, Fus2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
SEM6B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 3) | NC score | 0.017968 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 1.81305 (rank : 4) | NC score | 0.017902 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
IF36_HUMAN
|
||||||
| θ value | 1.81305 (rank : 5) | NC score | 0.068607 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60228, O43902, Q64058, Q64059, Q64252 | Gene names | EIF3S6, INT6 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 6 (eIF-3 p48) (eIF3e) (Viral integration site protein INT-6 homolog). | |||||
|
IF36_MOUSE
|
||||||
| θ value | 1.81305 (rank : 6) | NC score | 0.068607 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60229, O43902, Q64058, Q64059, Q64252 | Gene names | Eif3s6, Int6 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 6 (eIF-3 p48) (eIF3e) (Mammary tumor-associated protein INT-6) (Viral integration site protein INT-6) (MMTV integration site 6). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.017323 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.013027 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.015801 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 10) | NC score | 0.015375 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.016813 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.036983 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.025592 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
TIE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.003326 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06806, Q811F4 | Gene names | Tie1, Tie, Tie-1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
CCD1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.027627 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.014849 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
FLNB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.011714 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
KLF4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.002456 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 694 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60793, P70421 | Gene names | Klf4, Ezf, Gklf, Zie | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Gut-enriched krueppel-like factor) (Epithelial zinc-finger protein EZF). | |||||
|
LRFN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.006067 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BLY3 | Gene names | Lrfn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 3 precursor. | |||||
|
LRP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.010470 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |
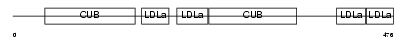
|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.031288 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
PCDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.004061 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
RASF7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.021856 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DD19 | Gene names | Rassf7 | |||
|
Domain Architecture |
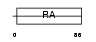
|
|||||
| Description | Ras association domain-containing protein 7. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.010309 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.012242 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
FAS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.014144 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
HSF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.014216 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0L1, Q9R0L2 | Gene names | Hsf4 | |||
|
Domain Architecture |
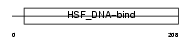
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (mHSF4). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.006214 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
UBQL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.012968 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
ZBTB9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.004583 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDC7, Q3U3B2, Q9CYQ0 | Gene names | Zbtb9 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 9. | |||||
|
NAT6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.20485e-131 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q93015, Q93014 | Gene names | NAT6, FUS2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
NAT6_MOUSE
|
||||||
| NC score | 0.968418 (rank : 2) | θ value | 2.01746e-92 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R123, Q8QZY5, Q8R014, Q9ERM0 | Gene names | Nat6, Fus2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
IF36_HUMAN
|
||||||
| NC score | 0.068607 (rank : 3) | θ value | 1.81305 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60228, O43902, Q64058, Q64059, Q64252 | Gene names | EIF3S6, INT6 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 6 (eIF-3 p48) (eIF3e) (Viral integration site protein INT-6 homolog). | |||||
|
IF36_MOUSE
|
||||||
| NC score | 0.068607 (rank : 4) | θ value | 1.81305 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60229, O43902, Q64058, Q64059, Q64252 | Gene names | Eif3s6, Int6 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 6 (eIF-3 p48) (eIF3e) (Mammary tumor-associated protein INT-6) (Viral integration site protein INT-6) (MMTV integration site 6). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.036983 (rank : 5) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.031288 (rank : 6) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
CCD1A_HUMAN
|
||||||
| NC score | 0.027627 (rank : 7) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.025592 (rank : 8) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
RASF7_MOUSE
|
||||||
| NC score | 0.021856 (rank : 9) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DD19 | Gene names | Rassf7 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 7. | |||||
|
SEM6B_MOUSE
|
||||||
| NC score | 0.017968 (rank : 10) | θ value | 1.06291 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.017902 (rank : 11) | θ value | 1.81305 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.017323 (rank : 12) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.016813 (rank : 13) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.015801 (rank : 14) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.015375 (rank : 15) | θ value | 4.03905 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.014849 (rank : 16) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
HSF4_MOUSE
|
||||||
| NC score | 0.014216 (rank : 17) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0L1, Q9R0L2 | Gene names | Hsf4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (mHSF4). | |||||
|
FAS_HUMAN
|
||||||
| NC score | 0.014144 (rank : 18) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.013027 (rank : 19) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
UBQL3_HUMAN
|
||||||
| NC score | 0.012968 (rank : 20) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.012242 (rank : 21) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
FLNB_HUMAN
|
||||||
| NC score | 0.011714 (rank : 22) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
LRP3_HUMAN
|
||||||
| NC score | 0.010470 (rank : 23) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.010309 (rank : 24) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.006214 (rank : 25) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
LRFN3_MOUSE
|
||||||
| NC score | 0.006067 (rank : 26) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BLY3 | Gene names | Lrfn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 3 precursor. | |||||
|
ZBTB9_MOUSE
|
||||||
| NC score | 0.004583 (rank : 27) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDC7, Q3U3B2, Q9CYQ0 | Gene names | Zbtb9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 9. | |||||
|
PCDC1_HUMAN
|
||||||
| NC score | 0.004061 (rank : 28) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
TIE1_MOUSE
|
||||||
| NC score | 0.003326 (rank : 29) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06806, Q811F4 | Gene names | Tie1, Tie, Tie-1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
KLF4_MOUSE
|
||||||
| NC score | 0.002456 (rank : 30) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 694 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60793, P70421 | Gene names | Klf4, Ezf, Gklf, Zie | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Gut-enriched krueppel-like factor) (Epithelial zinc-finger protein EZF). | |||||