Please be patient as the page loads
|
STAT6_HUMAN
|
||||||
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STAT6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
STAT6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.986300 (rank : 2) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
STA5B_HUMAN
|
||||||
| θ value | 2.13557e-126 (rank : 3) | NC score | 0.940930 (rank : 3) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51692, Q8WWS8 | Gene names | STAT5B | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5A_MOUSE
|
||||||
| θ value | 2.78914e-126 (rank : 4) | NC score | 0.938077 (rank : 4) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
STA5A_HUMAN
|
||||||
| θ value | 3.64273e-126 (rank : 5) | NC score | 0.936135 (rank : 6) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42229 | Gene names | STAT5A, STAT5 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A. | |||||
|
STA5B_MOUSE
|
||||||
| θ value | 1.80787e-125 (rank : 6) | NC score | 0.938051 (rank : 5) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P42232, Q541Q5, Q60804, Q8K3Q1, Q9JKM1 | Gene names | Stat5b | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STAT4_MOUSE
|
||||||
| θ value | 1.41344e-53 (rank : 7) | NC score | 0.867572 (rank : 7) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT4_HUMAN
|
||||||
| θ value | 1.84601e-53 (rank : 8) | NC score | 0.863068 (rank : 8) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT3_HUMAN
|
||||||
| θ value | 5.7518e-47 (rank : 9) | NC score | 0.850281 (rank : 9) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40763, O14916, Q9BW54 | Gene names | STAT3, APRF | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT3_MOUSE
|
||||||
| θ value | 5.7518e-47 (rank : 10) | NC score | 0.850278 (rank : 10) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42227 | Gene names | Stat3, Aprf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT1_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 11) | NC score | 0.844496 (rank : 12) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
STAT2_HUMAN
|
||||||
| θ value | 8.59492e-43 (rank : 12) | NC score | 0.844845 (rank : 11) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
STAT1_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 13) | NC score | 0.840955 (rank : 13) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42225 | Gene names | Stat1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1. | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 4.00176e-32 (rank : 14) | NC score | 0.795069 (rank : 14) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.055981 (rank : 19) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.014336 (rank : 95) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.037034 (rank : 35) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.068006 (rank : 16) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
GPR64_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.019875 (rank : 76) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.053329 (rank : 21) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.054994 (rank : 20) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.025380 (rank : 58) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.030244 (rank : 47) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
NBEA_MOUSE
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.020035 (rank : 74) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
RUSC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.029371 (rank : 50) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.052889 (rank : 22) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.056248 (rank : 18) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.044519 (rank : 29) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.039216 (rank : 33) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.028833 (rank : 53) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.034643 (rank : 38) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.011332 (rank : 106) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.004207 (rank : 125) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.016616 (rank : 88) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.049087 (rank : 24) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.017216 (rank : 87) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.030838 (rank : 41) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.012851 (rank : 100) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
WASF3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.029367 (rank : 51) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.023009 (rank : 66) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.019620 (rank : 79) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.024789 (rank : 59) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
STRC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.030632 (rank : 45) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
AMELX_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.036981 (rank : 36) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.049140 (rank : 23) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.023371 (rank : 64) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.025602 (rank : 56) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.021008 (rank : 72) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.014723 (rank : 94) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
YTHD3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.019813 (rank : 78) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYK6, Q3UVI5, Q6NXJ6, Q6NXJ8, Q8BKB6 | Gene names | Ythdf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.018504 (rank : 82) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GLE1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.019918 (rank : 75) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
P85B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.045261 (rank : 28) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
P85B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.045780 (rank : 26) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.019491 (rank : 80) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.005098 (rank : 122) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
US6NL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.021791 (rank : 68) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
ALEX_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.029895 (rank : 48) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.023622 (rank : 62) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.042776 (rank : 31) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.030662 (rank : 43) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.023733 (rank : 61) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
PRCC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.030443 (rank : 46) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.028678 (rank : 54) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.017682 (rank : 85) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SYAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.032600 (rank : 40) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96A49, Q96C60, Q96JQ6, Q96T20 | Gene names | SYAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapse-associated protein 1. | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.019854 (rank : 77) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
US6NL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.021434 (rank : 69) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
AMELX_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.030644 (rank : 44) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63277, P45559, P70592, Q61293 | Gene names | Amelx, Amel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amelogenin, X isoform precursor (Leucine-rich amelogenin peptide) (LRAP). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.038481 (rank : 34) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.005173 (rank : 121) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.045660 (rank : 27) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GP176_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.003263 (rank : 128) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
PRR8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.029085 (rank : 52) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
SON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.034731 (rank : 37) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.003395 (rank : 127) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.018049 (rank : 83) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
UACA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.013808 (rank : 98) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.023232 (rank : 65) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.009576 (rank : 112) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.015865 (rank : 91) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.019053 (rank : 81) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.025653 (rank : 55) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.015674 (rank : 92) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.013591 (rank : 99) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.003548 (rank : 126) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.029377 (rank : 49) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
KI21B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.006170 (rank : 120) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
KI21B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.006178 (rank : 118) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.002525 (rank : 130) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.014195 (rank : 96) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
TOX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.010135 (rank : 111) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.030765 (rank : 42) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.033829 (rank : 39) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.021117 (rank : 71) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
BARH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.001235 (rank : 131) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VIB5, Q66L43 | Gene names | Barhl2, Barh1, Mbh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 2 homeobox protein (Bar-class homeodomain protein MBH1) (Homeobox protein B-H1). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.023895 (rank : 60) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.017813 (rank : 84) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CDC6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.009429 (rank : 113) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99741, Q8TB30 | Gene names | CDC6, CDC18L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)) (HsCDC6) (HsCDC18). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.014747 (rank : 93) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
HSH2D_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.059584 (rank : 17) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.014050 (rank : 97) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LIPB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.010652 (rank : 109) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.000058 (rank : 132) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.008463 (rank : 114) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
PRD14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | -0.002965 (rank : 133) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZV8, Q86UX9 | Gene names | PRDM14 | |||
|
Domain Architecture |
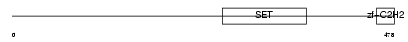
|
|||||
| Description | PR domain zinc finger protein 14 (PR domain-containing protein 14). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.025449 (rank : 57) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.022812 (rank : 67) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.023421 (rank : 63) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.016590 (rank : 89) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SYAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.020875 (rank : 73) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D5V6, Q3UI67, Q9D870 | Gene names | Syap1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapse-associated protein 1. | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.010792 (rank : 107) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.010684 (rank : 108) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TPO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.021383 (rank : 70) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
WNT9A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.002716 (rank : 129) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14904, Q5VWU0, Q96S50 | Gene names | WNT9A, WNT14 | |||
|
Domain Architecture |
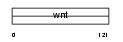
|
|||||
| Description | Protein Wnt-9a precursor (Wnt-14). | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.010419 (rank : 110) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CN092_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.011565 (rank : 104) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN102_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.008240 (rank : 115) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80XC6, Q8R3D7, Q99LT9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf102 homolog. | |||||
|
CTNA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.004305 (rank : 124) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q65CL1, Q3UQ61, Q8C0N3 | Gene names | Ctnna3, Catna3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Catenin alpha-3 (Alpha T-catenin) (Cadherin-associated protein). | |||||
|
CU070_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.011814 (rank : 103) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSI2 | Gene names | C21orf70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf70. | |||||
|
FAS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.006177 (rank : 119) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.016506 (rank : 90) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
INVO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.012095 (rank : 101) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
LPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.006449 (rank : 116) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.006193 (rank : 117) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
P55G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.043136 (rank : 30) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P55G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.048264 (rank : 25) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.017460 (rank : 86) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.004322 (rank : 123) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.011987 (rank : 102) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.039793 (rank : 32) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.011487 (rank : 105) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SH22A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.081332 (rank : 15) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
STAT6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
STAT6_MOUSE
|
||||||
| NC score | 0.986300 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
STA5B_HUMAN
|
||||||
| NC score | 0.940930 (rank : 3) | θ value | 2.13557e-126 (rank : 3) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51692, Q8WWS8 | Gene names | STAT5B | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5A_MOUSE
|
||||||
| NC score | 0.938077 (rank : 4) | θ value | 2.78914e-126 (rank : 4) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
STA5B_MOUSE
|
||||||
| NC score | 0.938051 (rank : 5) | θ value | 1.80787e-125 (rank : 6) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P42232, Q541Q5, Q60804, Q8K3Q1, Q9JKM1 | Gene names | Stat5b | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5A_HUMAN
|
||||||
| NC score | 0.936135 (rank : 6) | θ value | 3.64273e-126 (rank : 5) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42229 | Gene names | STAT5A, STAT5 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A. | |||||
|
STAT4_MOUSE
|
||||||
| NC score | 0.867572 (rank : 7) | θ value | 1.41344e-53 (rank : 7) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT4_HUMAN
|
||||||
| NC score | 0.863068 (rank : 8) | θ value | 1.84601e-53 (rank : 8) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT3_HUMAN
|
||||||
| NC score | 0.850281 (rank : 9) | θ value | 5.7518e-47 (rank : 9) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40763, O14916, Q9BW54 | Gene names | STAT3, APRF | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT3_MOUSE
|
||||||
| NC score | 0.850278 (rank : 10) | θ value | 5.7518e-47 (rank : 10) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42227 | Gene names | Stat3, Aprf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT2_HUMAN
|
||||||
| NC score | 0.844845 (rank : 11) | θ value | 8.59492e-43 (rank : 12) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
STAT1_HUMAN
|
||||||
| NC score | 0.844496 (rank : 12) | θ value | 2.95404e-43 (rank : 11) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
STAT1_MOUSE
|
||||||
| NC score | 0.840955 (rank : 13) | θ value | 2.11701e-41 (rank : 13) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42225 | Gene names | Stat1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1. | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.795069 (rank : 14) | θ value | 4.00176e-32 (rank : 14) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
SH22A_HUMAN
|
||||||
| NC score | 0.081332 (rank : 15) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.068006 (rank : 16) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
HSH2D_MOUSE
|
||||||
| NC score | 0.059584 (rank : 17) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.056248 (rank : 18) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.055981 (rank : 19) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.054994 (rank : 20) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.053329 (rank : 21) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.052889 (rank : 22) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.049140 (rank : 23) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.049087 (rank : 24) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
P55G_MOUSE
|
||||||
| NC score | 0.048264 (rank : 25) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.045780 (rank : 26) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.045660 (rank : 27) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.045261 (rank : 28) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.044519 (rank : 29) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
P55G_HUMAN
|
||||||
| NC score | 0.043136 (rank : 30) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.042776 (rank : 31) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.039793 (rank : 32) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.039216 (rank : 33) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.038481 (rank : 34) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.037034 (rank : 35) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
AMELX_HUMAN
|
||||||
| NC score | 0.036981 (rank : 36) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.034731 (rank : 37) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.034643 (rank : 38) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.033829 (rank : 39) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
SYAP1_HUMAN
|
||||||
| NC score | 0.032600 (rank : 40) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96A49, Q96C60, Q96JQ6, Q96T20 | Gene names | SYAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapse-associated protein 1. | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.030838 (rank : 41) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.030765 (rank : 42) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.030662 (rank : 43) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
AMELX_MOUSE
|
||||||
| NC score | 0.030644 (rank : 44) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63277, P45559, P70592, Q61293 | Gene names | Amelx, Amel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amelogenin, X isoform precursor (Leucine-rich amelogenin peptide) (LRAP). | |||||
|
STRC_HUMAN
|
||||||
| NC score | 0.030632 (rank : 45) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
PRCC_HUMAN
|
||||||
| NC score | 0.030443 (rank : 46) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.030244 (rank : 47) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.029895 (rank : 48) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.029377 (rank : 49) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
RUSC1_HUMAN
|
||||||
| NC score | 0.029371 (rank : 50) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
WASF3_HUMAN
|
||||||
| NC score | 0.029367 (rank : 51) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
PRR8_HUMAN
|
||||||
| NC score | 0.029085 (rank : 52) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.028833 (rank : 53) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.028678 (rank : 54) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.025653 (rank : 55) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.025602 (rank : 56) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.025449 (rank : 57) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.025380 (rank : 58) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.024789 (rank : 59) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.023895 (rank : 60) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.023733 (rank : 61) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.023622 (rank : 62) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.023421 (rank : 63) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.023371 (rank : 64) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.023232 (rank : 65) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.023009 (rank : 66) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.022812 (rank : 67) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
US6NL_MOUSE
|
||||||
| NC score | 0.021791 (rank : 68) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
US6NL_HUMAN
|
||||||
| NC score | 0.021434 (rank : 69) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.021383 (rank : 70) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.021117 (rank : 71) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.021008 (rank : 72) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SYAP1_MOUSE
|
||||||
| NC score | 0.020875 (rank : 73) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D5V6, Q3UI67, Q9D870 | Gene names | Syap1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapse-associated protein 1. | |||||
|
NBEA_MOUSE
|
||||||
| NC score | 0.020035 (rank : 74) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
GLE1_MOUSE
|
||||||
| NC score | 0.019918 (rank : 75) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
GPR64_HUMAN
|
||||||
| NC score | 0.019875 (rank : 76) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.019854 (rank : 77) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
YTHD3_MOUSE
|
||||||
| NC score | 0.019813 (rank : 78) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYK6, Q3UVI5, Q6NXJ6, Q6NXJ8, Q8BKB6 | Gene names | Ythdf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.019620 (rank : 79) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.019491 (rank : 80) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.019053 (rank : 81) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.018504 (rank : 82) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.018049 (rank : 83) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.017813 (rank : 84) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.017682 (rank : 85) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.017460 (rank : 86) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.017216 (rank : 87) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.016616 (rank : 88) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.016590 (rank : 89) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.016506 (rank : 90) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.015865 (rank : 91) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.015674 (rank : 92) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.014747 (rank : 93) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.014723 (rank : 94) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.014336 (rank : 95) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.014195 (rank : 96) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.014050 (rank : 97) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.013808 (rank : 98) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.013591 (rank : 99) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.012851 (rank : 100) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.012095 (rank : 101) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.011987 (rank : 102) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
CU070_HUMAN
|
||||||
| NC score | 0.011814 (rank : 103) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSI2 | Gene names | C21orf70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf70. | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.011565 (rank : 104) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.011487 (rank : 105) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.011332 (rank : 106) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.010792 (rank : 107) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.010684 (rank : 108) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
LIPB1_MOUSE
|
||||||
| NC score | 0.010652 (rank : 109) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.010419 (rank : 110) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
TOX_HUMAN
|
||||||
| NC score | 0.010135 (rank : 111) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.009576 (rank : 112) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
CDC6_HUMAN
|
||||||
| NC score | 0.009429 (rank : 113) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99741, Q8TB30 | Gene names | CDC6, CDC18L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)) (HsCDC6) (HsCDC18). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.008463 (rank : 114) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
CN102_MOUSE
|
||||||
| NC score | 0.008240 (rank : 115) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80XC6, Q8R3D7, Q99LT9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf102 homolog. | |||||
|
LPP_MOUSE
|
||||||
| NC score | 0.006449 (rank : 116) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.006193 (rank : 117) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
KI21B_MOUSE
|
||||||
| NC score | 0.006178 (rank : 118) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
FAS_HUMAN
|
||||||
| NC score | 0.006177 (rank : 119) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
KI21B_HUMAN
|
||||||
| NC score | 0.006170 (rank : 120) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.005173 (rank : 121) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.005098 (rank : 122) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.004322 (rank : 123) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
CTNA3_MOUSE
|
||||||
| NC score | 0.004305 (rank : 124) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q65CL1, Q3UQ61, Q8C0N3 | Gene names | Ctnna3, Catna3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Catenin alpha-3 (Alpha T-catenin) (Cadherin-associated protein). | |||||
|
MK07_HUMAN
|
||||||
| NC score | 0.004207 (rank : 125) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.003548 (rank : 126) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.003395 (rank : 127) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
GP176_MOUSE
|
||||||
| NC score | 0.003263 (rank : 128) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
WNT9A_HUMAN
|
||||||
| NC score | 0.002716 (rank : 129) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14904, Q5VWU0, Q96S50 | Gene names | WNT9A, WNT14 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-9a precursor (Wnt-14). | |||||
|
PCD15_HUMAN
|
||||||
| NC score | 0.002525 (rank : 130) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
BARH2_MOUSE
|
||||||
| NC score | 0.001235 (rank : 131) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VIB5, Q66L43 | Gene names | Barhl2, Barh1, Mbh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 2 homeobox protein (Bar-class homeodomain protein MBH1) (Homeobox protein B-H1). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | 0.000058 (rank : 132) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
PRD14_HUMAN
|
||||||
| NC score | -0.002965 (rank : 133) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZV8, Q86UX9 | Gene names | PRDM14 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 14 (PR domain-containing protein 14). | |||||