Please be patient as the page loads
|
STA5B_HUMAN
|
||||||
| SwissProt Accessions | P51692, Q8WWS8 | Gene names | STAT5B | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STA5A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994214 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P42229 | Gene names | STAT5A, STAT5 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A. | |||||
|
STA5A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994843 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
STA5B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P51692, Q8WWS8 | Gene names | STAT5B | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.997104 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P42232, Q541Q5, Q60804, Q8K3Q1, Q9JKM1 | Gene names | Stat5b | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STAT6_HUMAN
|
||||||
| θ value | 2.13557e-126 (rank : 5) | NC score | 0.940930 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
STAT6_MOUSE
|
||||||
| θ value | 1.05987e-125 (rank : 6) | NC score | 0.947292 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
STAT4_HUMAN
|
||||||
| θ value | 2.64101e-84 (rank : 7) | NC score | 0.898109 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT3_HUMAN
|
||||||
| θ value | 4.50487e-84 (rank : 8) | NC score | 0.892413 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P40763, O14916, Q9BW54 | Gene names | STAT3, APRF | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT3_MOUSE
|
||||||
| θ value | 4.50487e-84 (rank : 9) | NC score | 0.892411 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P42227 | Gene names | Stat3, Aprf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT4_MOUSE
|
||||||
| θ value | 5.50685e-82 (rank : 10) | NC score | 0.902819 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT1_HUMAN
|
||||||
| θ value | 3.69378e-78 (rank : 11) | NC score | 0.889328 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
STAT1_MOUSE
|
||||||
| θ value | 6.96618e-77 (rank : 12) | NC score | 0.886631 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P42225 | Gene names | Stat1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1. | |||||
|
STAT2_HUMAN
|
||||||
| θ value | 4.39379e-55 (rank : 13) | NC score | 0.875535 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 2.3352e-48 (rank : 14) | NC score | 0.821279 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 15) | NC score | 0.085881 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 16) | NC score | 0.071748 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 17) | NC score | 0.079062 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.068214 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.075103 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.055824 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.074409 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.069848 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.065578 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.033457 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
SH22A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.117191 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.061619 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.067501 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
APOE_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.053035 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08226 | Gene names | Apoe | |||
|
Domain Architecture |
|
|||||
| Description | Apolipoprotein E precursor (Apo-E). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.061999 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.062933 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.033836 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
HAPIP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.046067 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
HSH2D_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.071696 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JZ2, Q6ZNG7 | Gene names | HSH2D, ALX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
P55G_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.071821 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P55G_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.077820 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.061767 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.047900 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.059694 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.047643 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.057368 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.040955 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.058262 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.020172 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.045588 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.061905 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.058217 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.055590 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CHIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.023066 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.051683 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
HSH2D_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.085155 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.078894 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.070894 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.063210 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.036277 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
CHIN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.020450 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.054050 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.055203 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
K1683_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.043180 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
SH3K1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.011746 (rank : 109) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96B97, Q8IWX6, Q8IX98, Q96RN4, Q9NYR0 | Gene names | SH3KBP1, CIN85 | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (Cbl-interacting protein of 85 kDa) (Human Src-family kinase-binding protein 1) (HSB-1) (CD2-binding protein 3) (CD2BP3). | |||||
|
DNMBP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.016763 (rank : 106) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
K22E_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.016422 (rank : 107) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.035567 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.041329 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.032577 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
P85A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.056807 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.055500 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
PPT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.027418 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMR5, O14799, Q0P6K0, Q5JP13, Q5JP14, Q5JQF0, Q5SSX4, Q5SSX5, Q5SSX6, Q5STJ4, Q5STJ5, Q5STJ6, Q6FI80, Q99945 | Gene names | PPT2 | |||
|
Domain Architecture |
|
|||||
| Description | Lysosomal thioesterase PPT2 precursor (EC 3.1.2.-) (PPT-2) (S- thioesterase G14). | |||||
|
CLCN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.005670 (rank : 110) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0A1, Q9WUJ9 | Gene names | Clcn2, Clc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 2 (ClC-2). | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.057102 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.053568 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.049659 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.029751 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.023660 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.039221 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.032056 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
P85B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.067054 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
RABE2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.051227 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.047565 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.042085 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
FOXP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.030504 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15409, Q6ZND1, Q75MJ3, Q8IZE0, Q8N0W2, Q8N6B7, Q8N6B8, Q8NFQ1, Q8NFQ2, Q8NFQ3, Q8NFQ4, Q8TD74 | Gene names | FOXP2, CAGH44, TNRC10 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P2 (CAG repeat protein 44) (Trinucleotide repeat- containing gene 10 protein). | |||||
|
FOXP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.031259 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P58463, Q6PD37, Q8C4F0, Q8R441 | Gene names | Foxp2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P2. | |||||
|
P85B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.067170 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PPT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.017829 (rank : 105) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35448, Q80WP5 | Gene names | Ppt2 | |||
|
Domain Architecture |
|
|||||
| Description | Lysosomal thioesterase PPT2 precursor (EC 3.1.2.-) (PPT-2). | |||||
|
SOCS4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.036890 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXH5 | Gene names | SOCS4, SOCS7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
SOCS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.039255 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZA6 | Gene names | Socs4, Socs7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.012574 (rank : 108) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
APOE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.030943 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P02649, Q9P2S4 | Gene names | APOE | |||
|
Domain Architecture |
|
|||||
| Description | Apolipoprotein E precursor (Apo-E). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.045847 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.051920 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.048838 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RBM19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.002291 (rank : 112) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
REST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.043893 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SRMS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.001410 (rank : 113) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |
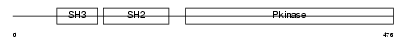
|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.052893 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.003730 (rank : 111) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.042650 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.047288 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.020147 (rank : 104) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.036909 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
SH22A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.063770 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
SOCS5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.025890 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75159, Q8IYZ4 | Gene names | SOCS5, CIS6, CISH5, CISH6, KIAA0671 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5) (Cytokine-inducible SH2 protein 6) (CIS-6). | |||||
|
SOCS5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.026110 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54928 | Gene names | Socs5, Cish5 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5). | |||||
|
TUFT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.046442 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
CCD11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.059612 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CCD27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.064166 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CCD27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.053569 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CF060_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.063225 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.050949 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.052205 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.055210 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.055773 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
REST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052497 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.055822 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
STA5B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P51692, Q8WWS8 | Gene names | STAT5B | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5B_MOUSE
|
||||||
| NC score | 0.997104 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P42232, Q541Q5, Q60804, Q8K3Q1, Q9JKM1 | Gene names | Stat5b | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5A_MOUSE
|
||||||
| NC score | 0.994843 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
STA5A_HUMAN
|
||||||
| NC score | 0.994214 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P42229 | Gene names | STAT5A, STAT5 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A. | |||||
|
STAT6_MOUSE
|
||||||
| NC score | 0.947292 (rank : 5) | θ value | 1.05987e-125 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
STAT6_HUMAN
|
||||||
| NC score | 0.940930 (rank : 6) | θ value | 2.13557e-126 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
STAT4_MOUSE
|
||||||
| NC score | 0.902819 (rank : 7) | θ value | 5.50685e-82 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT4_HUMAN
|
||||||
| NC score | 0.898109 (rank : 8) | θ value | 2.64101e-84 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT3_HUMAN
|
||||||
| NC score | 0.892413 (rank : 9) | θ value | 4.50487e-84 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P40763, O14916, Q9BW54 | Gene names | STAT3, APRF | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT3_MOUSE
|
||||||
| NC score | 0.892411 (rank : 10) | θ value | 4.50487e-84 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P42227 | Gene names | Stat3, Aprf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT1_HUMAN
|
||||||
| NC score | 0.889328 (rank : 11) | θ value | 3.69378e-78 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
STAT1_MOUSE
|
||||||
| NC score | 0.886631 (rank : 12) | θ value | 6.96618e-77 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P42225 | Gene names | Stat1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1. | |||||
|
STAT2_HUMAN
|
||||||
| NC score | 0.875535 (rank : 13) | θ value | 4.39379e-55 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.821279 (rank : 14) | θ value | 2.3352e-48 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
SH22A_HUMAN
|
||||||
| NC score | 0.117191 (rank : 15) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.085881 (rank : 16) | θ value | 7.1131e-05 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HSH2D_MOUSE
|
||||||
| NC score | 0.085155 (rank : 17) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.079062 (rank : 18) | θ value | 0.00665767 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.078894 (rank : 19) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
P55G_MOUSE
|
||||||
| NC score | 0.077820 (rank : 20) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.075103 (rank : 21) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.074409 (rank : 22) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
P55G_HUMAN
|
||||||
| NC score | 0.071821 (rank : 23) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.071748 (rank : 24) | θ value | 0.00509761 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
HSH2D_HUMAN
|
||||||
| NC score | 0.071696 (rank : 25) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JZ2, Q6ZNG7 | Gene names | HSH2D, ALX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.070894 (rank : 26) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.069848 (rank : 27) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.068214 (rank : 28) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.067501 (rank : 29) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.067170 (rank : 30) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.067054 (rank : 31) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.065578 (rank : 32) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CCD27_HUMAN
|
||||||
| NC score | 0.064166 (rank : 33) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
SH22A_MOUSE
|
||||||
| NC score | 0.063770 (rank : 34) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.063225 (rank : 35) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.063210 (rank : 36) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.062933 (rank : 37) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.061999 (rank : 38) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.061905 (rank : 39) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.061767 (rank : 40) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.061619 (rank : 41) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.059694 (rank : 42) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.059612 (rank : 43) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.058262 (rank : 44) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.058217 (rank : 45) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.057368 (rank : 46) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.057102 (rank : 47) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.056807 (rank : 48) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.055824 (rank : 49) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.055822 (rank : 50) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.055773 (rank : 51) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.055590 (rank : 52) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.055500 (rank : 53) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.055210 (rank : 54) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.055203 (rank : 55) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.054050 (rank : 56) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CCD27_MOUSE
|
||||||
| NC score | 0.053569 (rank : 57) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.053568 (rank : 58) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
APOE_MOUSE
|
||||||
| NC score | 0.053035 (rank : 59) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08226 | Gene names | Apoe | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein E precursor (Apo-E). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.052893 (rank : 60) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.052497 (rank : 61) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.052205 (rank : 62) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.051920 (rank : 63) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.051683 (rank : 64) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
RABE2_MOUSE
|
||||||
| NC score | 0.051227 (rank : 65) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.050949 (rank : 66) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.049659 (rank : 67) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.048838 (rank : 68) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.047900 (rank : 69) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.047643 (rank : 70) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.047565 (rank : 71) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.047288 (rank : 72) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
TUFT1_HUMAN
|
||||||
| NC score | 0.046442 (rank : 73) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
HAPIP_HUMAN
|
||||||
| NC score | 0.046067 (rank : 74) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.045847 (rank : 75) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.045588 (rank : 76) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.043893 (rank : 77) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.043180 (rank : 78) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.042650 (rank : 79) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.042085 (rank : 80) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.041329 (rank : 81) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.040955 (rank : 82) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
SOCS4_MOUSE
|
||||||
| NC score | 0.039255 (rank : 83) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZA6 | Gene names | Socs4, Socs7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.039221 (rank : 84) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.036909 (rank : 85) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
SOCS4_HUMAN
|
||||||
| NC score | 0.036890 (rank : 86) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXH5 | Gene names | SOCS4, SOCS7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.036277 (rank : 87) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.035567 (rank : 88) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.033836 (rank : 89) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.033457 (rank : 90) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.032577 (rank : 91) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.032056 (rank : 92) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
FOXP2_MOUSE
|
||||||
| NC score | 0.031259 (rank : 93) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P58463, Q6PD37, Q8C4F0, Q8R441 | Gene names | Foxp2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P2. | |||||
|
APOE_HUMAN
|
||||||
| NC score | 0.030943 (rank : 94) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P02649, Q9P2S4 | Gene names | APOE | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein E precursor (Apo-E). | |||||
|
FOXP2_HUMAN
|
||||||
| NC score | 0.030504 (rank : 95) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15409, Q6ZND1, Q75MJ3, Q8IZE0, Q8N0W2, Q8N6B7, Q8N6B8, Q8NFQ1, Q8NFQ2, Q8NFQ3, Q8NFQ4, Q8TD74 | Gene names | FOXP2, CAGH44, TNRC10 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P2 (CAG repeat protein 44) (Trinucleotide repeat- containing gene 10 protein). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.029751 (rank : 96) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
PPT2_HUMAN
|
||||||
| NC score | 0.027418 (rank : 97) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMR5, O14799, Q0P6K0, Q5JP13, Q5JP14, Q5JQF0, Q5SSX4, Q5SSX5, Q5SSX6, Q5STJ4, Q5STJ5, Q5STJ6, Q6FI80, Q99945 | Gene names | PPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal thioesterase PPT2 precursor (EC 3.1.2.-) (PPT-2) (S- thioesterase G14). | |||||
|
SOCS5_MOUSE
|
||||||
| NC score | 0.026110 (rank : 98) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54928 | Gene names | Socs5, Cish5 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5). | |||||
|
SOCS5_HUMAN
|
||||||
| NC score | 0.025890 (rank : 99) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75159, Q8IYZ4 | Gene names | SOCS5, CIS6, CISH5, CISH6, KIAA0671 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5) (Cytokine-inducible SH2 protein 6) (CIS-6). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.023660 (rank : 100) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
CHIN_HUMAN
|
||||||
| NC score | 0.023066 (rank : 101) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIN_MOUSE
|
||||||
| NC score | 0.020450 (rank : 102) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.020172 (rank : 103) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.020147 (rank : 104) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
PPT2_MOUSE
|
||||||
| NC score | 0.017829 (rank : 105) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35448, Q80WP5 | Gene names | Ppt2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal thioesterase PPT2 precursor (EC 3.1.2.-) (PPT-2). | |||||
|
DNMBP_HUMAN
|
||||||
| NC score | 0.016763 (rank : 106) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
K22E_HUMAN
|
||||||
| NC score | 0.016422 (rank : 107) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.012574 (rank : 108) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SH3K1_HUMAN
|
||||||
| NC score | 0.011746 (rank : 109) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96B97, Q8IWX6, Q8IX98, Q96RN4, Q9NYR0 | Gene names | SH3KBP1, CIN85 | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (Cbl-interacting protein of 85 kDa) (Human Src-family kinase-binding protein 1) (HSB-1) (CD2-binding protein 3) (CD2BP3). | |||||
|
CLCN2_MOUSE
|
||||||
| NC score | 0.005670 (rank : 110) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0A1, Q9WUJ9 | Gene names | Clcn2, Clc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 2 (ClC-2). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.003730 (rank : 111) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
RBM19_HUMAN
|
||||||
| NC score | 0.002291 (rank : 112) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
SRMS_HUMAN
|
||||||
| NC score | 0.001410 (rank : 113) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||