Please be patient as the page loads
|
STAT2_MOUSE
|
||||||
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STAT2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.945423 (rank : 2) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 175 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
STAT1_HUMAN
|
||||||
| θ value | 2.77624e-142 (rank : 3) | NC score | 0.914295 (rank : 5) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
STAT1_MOUSE
|
||||||
| θ value | 1.98959e-140 (rank : 4) | NC score | 0.913431 (rank : 6) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P42225 | Gene names | Stat1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1. | |||||
|
STAT4_HUMAN
|
||||||
| θ value | 1.80368e-133 (rank : 5) | NC score | 0.915475 (rank : 4) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT3_HUMAN
|
||||||
| θ value | 2.20485e-131 (rank : 6) | NC score | 0.907802 (rank : 7) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P40763, O14916, Q9BW54 | Gene names | STAT3, APRF | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT3_MOUSE
|
||||||
| θ value | 2.20485e-131 (rank : 7) | NC score | 0.907801 (rank : 8) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P42227 | Gene names | Stat3, Aprf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT4_MOUSE
|
||||||
| θ value | 1.29561e-123 (rank : 8) | NC score | 0.918194 (rank : 3) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STA5B_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 9) | NC score | 0.821279 (rank : 9) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51692, Q8WWS8 | Gene names | STAT5B | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5B_MOUSE
|
||||||
| θ value | 6.7944e-48 (rank : 10) | NC score | 0.818896 (rank : 10) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42232, Q541Q5, Q60804, Q8K3Q1, Q9JKM1 | Gene names | Stat5b | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5A_MOUSE
|
||||||
| θ value | 7.51208e-47 (rank : 11) | NC score | 0.816370 (rank : 11) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
STA5A_HUMAN
|
||||||
| θ value | 2.18568e-46 (rank : 12) | NC score | 0.813506 (rank : 12) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42229 | Gene names | STAT5A, STAT5 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A. | |||||
|
STAT6_HUMAN
|
||||||
| θ value | 4.00176e-32 (rank : 13) | NC score | 0.795069 (rank : 13) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
STAT6_MOUSE
|
||||||
| θ value | 7.54701e-31 (rank : 14) | NC score | 0.793713 (rank : 14) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 15) | NC score | 0.105414 (rank : 20) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 16) | NC score | 0.102707 (rank : 22) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
GNDS_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 17) | NC score | 0.101772 (rank : 23) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 18) | NC score | 0.101343 (rank : 24) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 19) | NC score | 0.102960 (rank : 21) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 20) | NC score | 0.107773 (rank : 19) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 21) | NC score | 0.057242 (rank : 56) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
TACC3_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 22) | NC score | 0.046853 (rank : 72) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 23) | NC score | 0.038518 (rank : 91) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 24) | NC score | 0.060073 (rank : 49) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 25) | NC score | 0.066933 (rank : 39) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 26) | NC score | 0.079005 (rank : 30) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
MOT8_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 27) | NC score | 0.058825 (rank : 54) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 28) | NC score | 0.055834 (rank : 57) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 29) | NC score | 0.016816 (rank : 135) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 30) | NC score | 0.046412 (rank : 74) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PHLA1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 31) | NC score | 0.077292 (rank : 31) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
PRAX_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 32) | NC score | 0.113241 (rank : 16) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 33) | NC score | 0.108404 (rank : 18) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 34) | NC score | 0.062209 (rank : 47) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 35) | NC score | 0.045315 (rank : 76) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 36) | NC score | 0.020784 (rank : 122) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 37) | NC score | 0.065688 (rank : 40) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 38) | NC score | 0.120915 (rank : 15) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 39) | NC score | 0.028870 (rank : 106) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 40) | NC score | 0.071009 (rank : 36) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
PHLA1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 41) | NC score | 0.074677 (rank : 34) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 42) | NC score | 0.096439 (rank : 25) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ABL2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 43) | NC score | 0.011642 (rank : 151) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 44) | NC score | 0.054534 (rank : 60) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
SELV_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 45) | NC score | 0.065162 (rank : 41) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
MNT_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 46) | NC score | 0.067535 (rank : 37) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 47) | NC score | 0.059634 (rank : 51) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.038558 (rank : 89) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
WASF1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 49) | NC score | 0.040415 (rank : 84) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R5H6, Q91W51, Q9ERQ9 | Gene names | Wasf1, Wave1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 50) | NC score | 0.024381 (rank : 117) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 51) | NC score | 0.043879 (rank : 78) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
GGN_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 52) | NC score | 0.044071 (rank : 77) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
SF3A1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 53) | NC score | 0.059081 (rank : 53) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
WASF1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 54) | NC score | 0.041524 (rank : 82) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92558 | Gene names | WASF1, KIAA0269, SCAR1, WAVE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1) (Verprolin homology domain-containing protein 1). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 55) | NC score | 0.020331 (rank : 125) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 56) | NC score | 0.010913 (rank : 155) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 57) | NC score | 0.018998 (rank : 127) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 58) | NC score | 0.041596 (rank : 81) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 59) | NC score | 0.037393 (rank : 94) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.163984 (rank : 60) | NC score | 0.046845 (rank : 73) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 61) | NC score | 0.054709 (rank : 59) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 62) | NC score | 0.011097 (rank : 153) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.009698 (rank : 161) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 64) | NC score | 0.049990 (rank : 67) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.009795 (rank : 159) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 66) | NC score | 0.037653 (rank : 93) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 67) | NC score | 0.037658 (rank : 92) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | 0.279714 (rank : 68) | NC score | 0.063529 (rank : 43) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 0.279714 (rank : 69) | NC score | 0.037030 (rank : 95) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 70) | NC score | 0.006255 (rank : 174) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 71) | NC score | 0.032429 (rank : 100) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MOT8_HUMAN
|
||||||
| θ value | 0.279714 (rank : 72) | NC score | 0.041384 (rank : 83) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 0.365318 (rank : 73) | NC score | 0.028681 (rank : 109) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CASB_HUMAN
|
||||||
| θ value | 0.365318 (rank : 74) | NC score | 0.053405 (rank : 62) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05814, Q4VAZ9 | Gene names | CSN2, CASB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-casein precursor. | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 0.365318 (rank : 75) | NC score | 0.016495 (rank : 138) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.365318 (rank : 76) | NC score | 0.049629 (rank : 68) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 77) | NC score | 0.048816 (rank : 69) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
NTBP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 78) | NC score | 0.038522 (rank : 90) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T7V8, Q49A22, Q6P1P9, Q9HAE6, Q9Y350 | Gene names | SCYL1BP1, NTKLBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (hNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 0.365318 (rank : 79) | NC score | 0.050754 (rank : 64) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
CI079_HUMAN
|
||||||
| θ value | 0.47712 (rank : 80) | NC score | 0.039989 (rank : 86) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
FRAT1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 81) | NC score | 0.025970 (rank : 111) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
MTF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 82) | NC score | 0.000379 (rank : 188) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
PHC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 83) | NC score | 0.039031 (rank : 87) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 84) | NC score | 0.025206 (rank : 115) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.62314 (rank : 85) | NC score | 0.046974 (rank : 70) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
STA13_HUMAN
|
||||||
| θ value | 0.62314 (rank : 86) | NC score | 0.018421 (rank : 129) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 0.62314 (rank : 87) | NC score | 0.018647 (rank : 128) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | 0.034519 (rank : 98) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | 0.017960 (rank : 130) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.038692 (rank : 88) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
MORC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 91) | NC score | 0.025263 (rank : 114) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
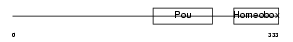
PO2F2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 92) | NC score | 0.015136 (rank : 144) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00196, Q00197, Q00198, Q00199, Q00200, Q00201, Q05882, Q61995, Q61996, Q64245 | Gene names | Pou2f2, Oct2, Otf2 | |||
|
Domain Architecture |
|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
RENBP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 93) | NC score | 0.031071 (rank : 104) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P82343, Q80Z67, Q9CXI1 | Gene names | Renbp | |||
|
Domain Architecture |
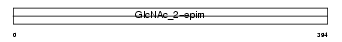
|
|||||
| Description | N-acylglucosamine 2-epimerase (EC 5.1.3.8) (GlcNAc 2-epimerase) (N- acetyl-D-glucosamine 2-epimerase) (AGE) (Renin-binding protein) (RnBP). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 94) | NC score | 0.028767 (rank : 108) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TF65_HUMAN
|
||||||
| θ value | 0.813845 (rank : 95) | NC score | 0.024390 (rank : 116) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 96) | NC score | 0.021285 (rank : 121) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 97) | NC score | 0.017745 (rank : 132) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 98) | NC score | 0.020508 (rank : 123) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
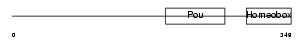
PO2F2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 99) | NC score | 0.016848 (rank : 134) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |
|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
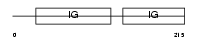
ROBO4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 100) | NC score | 0.011498 (rank : 152) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WZ75, Q8TEG1, Q96JV6, Q9H718, Q9NWJ8 | Gene names | ROBO4 | |||
|
Domain Architecture |
|
|||||
| Description | Roundabout homolog 4 precursor (Magic roundabout). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 101) | NC score | 0.023487 (rank : 118) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 102) | NC score | 0.009695 (rank : 162) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
MOR2B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 103) | NC score | 0.021387 (rank : 120) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5W4, Q8CG24 | Gene names | Morc2b, Tce6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2B (TCE6). | |||||
|
NTBP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 104) | NC score | 0.029364 (rank : 105) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
ZXDA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 105) | NC score | -0.001132 (rank : 189) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98168, Q9UJP7 | Gene names | ZXDA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDA. | |||||
|
P85B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 106) | NC score | 0.046266 (rank : 75) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 107) | NC score | 0.016583 (rank : 137) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 108) | NC score | 0.015850 (rank : 143) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
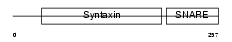
STX4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 109) | NC score | 0.007992 (rank : 166) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12846, Q15525 | Gene names | STX4, STX4A | |||
|
Domain Architecture |
|
|||||
| Description | Syntaxin-4 (NY-REN-31 antigen). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 110) | NC score | 0.015955 (rank : 142) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 111) | NC score | 0.025706 (rank : 113) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 112) | NC score | 0.109244 (rank : 17) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
UTP20_HUMAN
|
||||||
| θ value | 2.36792 (rank : 113) | NC score | 0.014066 (rank : 146) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.010214 (rank : 156) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.028783 (rank : 107) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.016619 (rank : 136) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.003250 (rank : 186) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
NRK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.003573 (rank : 185) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
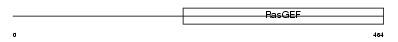
RGDSR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | 0.059728 (rank : 50) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZJ4 | Gene names | RGR | |||
|
Domain Architecture |
|
|||||
| Description | Ral-GDS-related protein (hRGR). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 120) | NC score | 0.009744 (rank : 160) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
AAK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.010021 (rank : 157) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.011854 (rank : 149) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
IRX3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.011692 (rank : 150) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78415, Q7Z4A4, Q7Z4A5, Q8IVC6 | Gene names | IRX3, IRXB1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.005653 (rank : 179) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.004099 (rank : 183) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
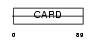
NOL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.031862 (rank : 101) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |
|
|||||
| Description | Nucleolar protein 3. | |||||
|
P55G_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.050539 (rank : 65) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P85B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.046885 (rank : 71) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
YTHD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.014060 (rank : 147) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z739, Q63Z37, Q659A3 | Gene names | YTHDF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
ZCHC7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.031294 (rank : 103) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
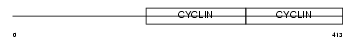
CCNB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.005842 (rank : 177) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
FYN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.005869 (rank : 176) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P06241 | Gene names | FYN | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn) (Protooncogene Syn) (SLK). | |||||
|
G3BP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.017312 (rank : 133) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13283 | Gene names | G3BP, G3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
GPR64_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.007608 (rank : 168) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
KLF11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | -0.001221 (rank : 190) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14901 | Gene names | KLF11, FKLF, TIEG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 11 (Transforming growth factor-beta-inducible early growth response protein 2) (TGFB-inducible early growth response protein 2) (TIEG-2). | |||||
|
KSR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.005482 (rank : 180) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6VAB6, Q8N775 | Gene names | KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-2 (hKSR2). | |||||
|
M3K14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.003573 (rank : 184) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99558, Q8IYN1 | Gene names | MAP3K14, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK) (HsNIK). | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.010008 (rank : 158) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.080128 (rank : 28) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.013339 (rank : 148) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
P85A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.041951 (rank : 80) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.034936 (rank : 97) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.014751 (rank : 145) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.004756 (rank : 182) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.025744 (rank : 112) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TSH3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.005752 (rank : 178) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
YTHD3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.016402 (rank : 139) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYK6, Q3UVI5, Q6NXJ6, Q6NXJ8, Q8BKB6 | Gene names | Ythdf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
EP400_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.026562 (rank : 110) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
FGR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.006406 (rank : 172) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P09769, Q9UIQ3 | Gene names | FGR, SRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FGR (EC 2.7.10.2) (P55-FGR) (C- FGR). | |||||
|
FGR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.006493 (rank : 171) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14234, Q61404 | Gene names | Fgr | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FGR (EC 2.7.10.2) (P55-FGR) (C- FGR). | |||||
|
FYN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.006293 (rank : 173) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39688 | Gene names | Fyn | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.032612 (rank : 99) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
K0460_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.017800 (rank : 131) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
KLF11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | -0.001430 (rank : 191) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EPF4 | Gene names | Klf11, Tieg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 11 (Transforming growth factor-beta-inducible early growth response protein 2) (TGFB-inducible early growth response protein 2) (TIEG-2). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.002678 (rank : 187) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
P55G_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.042695 (rank : 79) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.040186 (rank : 85) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.021528 (rank : 119) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SH22A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.091370 (rank : 27) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
SSH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.005155 (rank : 181) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
TBC10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.006207 (rank : 175) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXI6, O76053, Q543A2 | Gene names | TBC1D10A, EPI64, TBC1D10 | |||
|
Domain Architecture |
|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
TMLH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.008857 (rank : 163) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVH6 | Gene names | TMLHE, TMLH | |||
|
Domain Architecture |
|
|||||
| Description | Trimethyllysine dioxygenase, mitochondrial precursor (EC 1.14.11.8) (Epsilon-trimethyllysine 2-oxoglutarate dioxygenase) (TML-alpha- ketoglutarate dioxygenase) (TML hydroxylase) (TML dioxygenase) (TMLD). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.007230 (rank : 169) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
VPS28_HUMAN
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.016200 (rank : 140) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UK41 | Gene names | VPS28 | |||
|
Domain Architecture |
|
|||||
| Description | Vacuolar protein sorting-associated protein 28 homolog. | |||||
|
VPS28_MOUSE
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | 0.016115 (rank : 141) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1C8 | Gene names | Vps28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 28 homolog (Caspase- activated DNase inhibitor that interacts with ASK1) (CIIA). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 166) | NC score | 0.020055 (rank : 126) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WNK4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 167) | NC score | 0.008482 (rank : 164) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 6.88961 (rank : 168) | NC score | 0.031653 (rank : 102) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
CRF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.010984 (rank : 154) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06850 | Gene names | CRH | |||
|
Domain Architecture |

|
|||||
| Description | Corticoliberin precursor (Corticotropin-releasing factor) (CRF) (Corticotropin-releasing hormone). | |||||
|
G3PT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.006740 (rank : 170) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14556, O60823, Q6JTT9, Q9HCU6 | Gene names | GAPDHS, GAPD2, GAPDH2, GAPDS | |||
|
Domain Architecture |
|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
P66B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.007659 (rank : 167) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHR5, Q8C9Q3 | Gene names | Gatad2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 beta (p66/p68) (GATA zinc finger domain- containing protein 2B). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.034948 (rank : 96) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SH22A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.062963 (rank : 44) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.008210 (rank : 165) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.020410 (rank : 124) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.059345 (rank : 52) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.067258 (rank : 38) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
K1683_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.054424 (rank : 61) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.076160 (rank : 33) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.076187 (rank : 32) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.060307 (rank : 48) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.058296 (rank : 55) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.055618 (rank : 58) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.062225 (rank : 46) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.052179 (rank : 63) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.065058 (rank : 42) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.079264 (rank : 29) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.093547 (rank : 26) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.062541 (rank : 45) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.072264 (rank : 35) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.050412 (rank : 66) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 175 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
STAT2_HUMAN
|
||||||
| NC score | 0.945423 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
STAT4_MOUSE
|
||||||
| NC score | 0.918194 (rank : 3) | θ value | 1.29561e-123 (rank : 8) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT4_HUMAN
|
||||||
| NC score | 0.915475 (rank : 4) | θ value | 1.80368e-133 (rank : 5) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT1_HUMAN
|
||||||
| NC score | 0.914295 (rank : 5) | θ value | 2.77624e-142 (rank : 3) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
STAT1_MOUSE
|
||||||
| NC score | 0.913431 (rank : 6) | θ value | 1.98959e-140 (rank : 4) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P42225 | Gene names | Stat1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1. | |||||
|
STAT3_HUMAN
|
||||||
| NC score | 0.907802 (rank : 7) | θ value | 2.20485e-131 (rank : 6) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P40763, O14916, Q9BW54 | Gene names | STAT3, APRF | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT3_MOUSE
|
||||||
| NC score | 0.907801 (rank : 8) | θ value | 2.20485e-131 (rank : 7) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P42227 | Gene names | Stat3, Aprf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STA5B_HUMAN
|
||||||
| NC score | 0.821279 (rank : 9) | θ value | 2.3352e-48 (rank : 9) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51692, Q8WWS8 | Gene names | STAT5B | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5B_MOUSE
|
||||||
| NC score | 0.818896 (rank : 10) | θ value | 6.7944e-48 (rank : 10) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42232, Q541Q5, Q60804, Q8K3Q1, Q9JKM1 | Gene names | Stat5b | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5A_MOUSE
|
||||||
| NC score | 0.816370 (rank : 11) | θ value | 7.51208e-47 (rank : 11) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
STA5A_HUMAN
|
||||||
| NC score | 0.813506 (rank : 12) | θ value | 2.18568e-46 (rank : 12) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42229 | Gene names | STAT5A, STAT5 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A. | |||||
|
STAT6_HUMAN
|
||||||
| NC score | 0.795069 (rank : 13) | θ value | 4.00176e-32 (rank : 13) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
STAT6_MOUSE
|
||||||
| NC score | 0.793713 (rank : 14) | θ value | 7.54701e-31 (rank : 14) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.120915 (rank : 15) | θ value | 0.00869519 (rank : 38) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.113241 (rank : 16) | θ value | 0.00298849 (rank : 32) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.109244 (rank : 17) | θ value | 2.36792 (rank : 112) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.108404 (rank : 18) | θ value | 0.00298849 (rank : 33) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.107773 (rank : 19) | θ value | 5.44631e-05 (rank : 20) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.105414 (rank : 20) | θ value | 5.8054e-15 (rank : 15) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.102960 (rank : 21) | θ value | 1.80886e-08 (rank : 19) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.102707 (rank : 22) | θ value | 3.52202e-12 (rank : 16) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.101772 (rank : 23) | θ value | 1.06045e-08 (rank : 17) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.101343 (rank : 24) | θ value | 1.06045e-08 (rank : 18) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.096439 (rank : 25) | θ value | 0.0252991 (rank : 42) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.093547 (rank : 26) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SH22A_HUMAN
|
||||||
| NC score | 0.091370 (rank : 27) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.080128 (rank : 28) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.079264 (rank : 29) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.079005 (rank : 30) | θ value | 0.000786445 (rank : 26) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
PHLA1_MOUSE
|
||||||
| NC score | 0.077292 (rank : 31) | θ value | 0.00298849 (rank : 31) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.076187 (rank : 32) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.076160 (rank : 33) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PHLA1_HUMAN
|
||||||
| NC score | 0.074677 (rank : 34) | θ value | 0.0193708 (rank : 41) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.072264 (rank : 35) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.071009 (rank : 36) | θ value | 0.0193708 (rank : 40) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.067535 (rank : 37) | θ value | 0.0431538 (rank : 46) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.067258 (rank : 38) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.066933 (rank : 39) | θ value | 0.000602161 (rank : 25) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.065688 (rank : 40) | θ value | 0.00869519 (rank : 37) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.065162 (rank : 41) | θ value | 0.0330416 (rank : 45) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.065058 (rank : 42) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.063529 (rank : 43) | θ value | 0.279714 (rank : 68) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
SH22A_MOUSE
|
||||||
| NC score | 0.062963 (rank : 44) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.062541 (rank : 45) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.062225 (rank : 46) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.062209 (rank : 47) | θ value | 0.00390308 (rank : 34) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.060307 (rank : 48) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.060073 (rank : 49) | θ value | 0.000602161 (rank : 24) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
RGDSR_HUMAN
|
||||||
| NC score | 0.059728 (rank : 50) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZJ4 | Gene names | RGR | |||
|
Domain Architecture |

|
|||||
| Description | Ral-GDS-related protein (hRGR). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.059634 (rank : 51) | θ value | 0.0563607 (rank : 47) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.059345 (rank : 52) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
SF3A1_HUMAN
|
||||||
| NC score | 0.059081 (rank : 53) | θ value | 0.0736092 (rank : 53) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
MOT8_MOUSE
|
||||||
| NC score | 0.058825 (rank : 54) | θ value | 0.00134147 (rank : 27) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.058296 (rank : 55) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.057242 (rank : 56) | θ value | 0.000158464 (rank : 21) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.055834 (rank : 57) | θ value | 0.00175202 (rank : 28) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.055618 (rank : 58) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.054709 (rank : 59) | θ value | 0.163984 (rank : 61) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.054534 (rank : 60) | θ value | 0.0330416 (rank : 44) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.054424 (rank : 61) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
CASB_HUMAN
|
||||||
| NC score | 0.053405 (rank : 62) | θ value | 0.365318 (rank : 74) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05814, Q4VAZ9 | Gene names | CSN2, CASB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-casein precursor. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.052179 (rank : 63) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.050754 (rank : 64) | θ value | 0.365318 (rank : 79) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
P55G_MOUSE
|
||||||
| NC score | 0.050539 (rank : 65) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.050412 (rank : 66) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.049990 (rank : 67) | θ value | 0.21417 (rank : 64) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.049629 (rank : 68) | θ value | 0.365318 (rank : 76) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.048816 (rank : 69) | θ value | 0.365318 (rank : 77) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.046974 (rank : 70) | θ value | 0.62314 (rank : 85) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.046885 (rank : 71) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
TACC3_MOUSE
|
||||||
| NC score | 0.046853 (rank : 72) | θ value | 0.000158464 (rank : 22) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.046845 (rank : 73) | θ value | 0.163984 (rank : 60) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.046412 (rank : 74) | θ value | 0.00298849 (rank : 30) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.046266 (rank : 75) | θ value | 1.81305 (rank : 106) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.045315 (rank : 76) | θ value | 0.00665767 (rank : 35) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.044071 (rank : 77) | θ value | 0.0736092 (rank : 52) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.043879 (rank : 78) | θ value | 0.0736092 (rank : 51) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
P55G_HUMAN
|
||||||
| NC score | 0.042695 (rank : 79) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.041951 (rank : 80) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.041596 (rank : 81) | θ value | 0.0961366 (rank : 58) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
WASF1_HUMAN
|
||||||
| NC score | 0.041524 (rank : 82) | θ value | 0.0736092 (rank : 54) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92558 | Gene names | WASF1, KIAA0269, SCAR1, WAVE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1) (Verprolin homology domain-containing protein 1). | |||||
|
MOT8_HUMAN
|
||||||
| NC score | 0.041384 (rank : 83) | θ value | 0.279714 (rank : 72) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
WASF1_MOUSE
|
||||||
| NC score | 0.040415 (rank : 84) | θ value | 0.0563607 (rank : 49) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R5H6, Q91W51, Q9ERQ9 | Gene names | Wasf1, Wave1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1). | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.040186 (rank : 85) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.039989 (rank : 86) | θ value | 0.47712 (rank : 80) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
PHC1_HUMAN
|
||||||
| NC score | 0.039031 (rank : 87) | θ value | 0.47712 (rank : 83) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.038692 (rank : 88) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.038558 (rank : 89) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
NTBP1_HUMAN
|
||||||
| NC score | 0.038522 (rank : 90) | θ value | 0.365318 (rank : 78) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T7V8, Q49A22, Q6P1P9, Q9HAE6, Q9Y350 | Gene names | SCYL1BP1, NTKLBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (hNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.038518 (rank : 91) | θ value | 0.00035302 (rank : 23) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.037658 (rank : 92) | θ value | 0.279714 (rank : 67) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.037653 (rank : 93) | θ value | 0.279714 (rank : 66) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.037393 (rank : 94) | θ value | 0.163984 (rank : 59) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.037030 (rank : 95) | θ value | 0.279714 (rank : 69) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.034948 (rank : 96) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.034936 (rank : 97) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.034519 (rank : 98) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.032612 (rank : 99) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.032429 (rank : 100) | θ value | 0.279714 (rank : 71) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NOL3_MOUSE
|
||||||
| NC score | 0.031862 (rank : 101) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein 3. | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.031653 (rank : 102) | θ value | 6.88961 (rank : 168) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ZCHC7_HUMAN
|
||||||
| NC score | 0.031294 (rank : 103) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
RENBP_MOUSE
|
||||||
| NC score | 0.031071 (rank : 104) | θ value | 0.813845 (rank : 93) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P82343, Q80Z67, Q9CXI1 | Gene names | Renbp | |||
|
Domain Architecture |
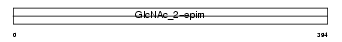
|
|||||
| Description | N-acylglucosamine 2-epimerase (EC 5.1.3.8) (GlcNAc 2-epimerase) (N- acetyl-D-glucosamine 2-epimerase) (AGE) (Renin-binding protein) (RnBP). | |||||
|
NTBP1_MOUSE
|
||||||
| NC score | 0.029364 (rank : 105) | θ value | 1.38821 (rank : 104) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.028870 (rank : 106) | θ value | 0.00869519 (rank : 39) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.028783 (rank : 107) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.028767 (rank : 108) | θ value | 0.813845 (rank : 94) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.028681 (rank : 109) | θ value | 0.365318 (rank : 73) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.026562 (rank : 110) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
FRAT1_HUMAN
|
||||||
| NC score | 0.025970 (rank : 111) | θ value | 0.47712 (rank : 81) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.025744 (rank : 112) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.025706 (rank : 113) | θ value | 2.36792 (rank : 111) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MORC2_HUMAN
|
||||||
| NC score | 0.025263 (rank : 114) | θ value | 0.813845 (rank : 91) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.025206 (rank : 115) | θ value | 0.62314 (rank : 84) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
TF65_HUMAN
|
||||||
| NC score | 0.024390 (rank : 116) | θ value | 0.813845 (rank : 95) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.024381 (rank : 117) | θ value | 0.0736092 (rank : 50) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.023487 (rank : 118) | θ value | 1.06291 (rank : 101) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.021528 (rank : 119) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
MOR2B_MOUSE
|
||||||
| NC score | 0.021387 (rank : 120) | θ value | 1.38821 (rank : 103) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5W4, Q8CG24 | Gene names | Morc2b, Tce6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2B (TCE6). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.021285 (rank : 121) | θ value | 1.06291 (rank : 96) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.020784 (rank : 122) | θ value | 0.00665767 (rank : 36) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.020508 (rank : 123) | θ value | 1.06291 (rank : 98) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.020410 (rank : 124) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.020331 (rank : 125) | θ value | 0.0961366 (rank : 55) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.020055 (rank : 126) | θ value | 6.88961 (rank : 166) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.018998 (rank : 127) | θ value | 0.0961366 (rank : 57) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.018647 (rank : 128) | θ value | 0.62314 (rank : 87) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.018421 (rank : 129) | θ value | 0.62314 (rank : 86) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.017960 (rank : 130) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.017800 (rank : 131) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.017745 (rank : 132) | θ value | 1.06291 (rank : 97) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
G3BP_HUMAN
|
||||||
| NC score | 0.017312 (rank : 133) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13283 | Gene names | G3BP, G3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
PO2F2_HUMAN
|
||||||
| NC score | 0.016848 (rank : 134) | θ value | 1.06291 (rank : 99) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.016816 (rank : 135) | θ value | 0.00228821 (rank : 29) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.016619 (rank : 136) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.016583 (rank : 137) | θ value | 1.81305 (rank : 107) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.016495 (rank : 138) | θ value | 0.365318 (rank : 75) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
YTHD3_MOUSE
|
||||||
| NC score | 0.016402 (rank : 139) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYK6, Q3UVI5, Q6NXJ6, Q6NXJ8, Q8BKB6 | Gene names | Ythdf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
VPS28_HUMAN
|
||||||
| NC score | 0.016200 (rank : 140) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UK41 | Gene names | VPS28 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 28 homolog. | |||||
|
VPS28_MOUSE
|
||||||
| NC score | 0.016115 (rank : 141) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1C8 | Gene names | Vps28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 28 homolog (Caspase- activated DNase inhibitor that interacts with ASK1) (CIIA). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.015955 (rank : 142) | θ value | 2.36792 (rank : 110) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.015850 (rank : 143) | θ value | 1.81305 (rank : 108) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
PO2F2_MOUSE
|
||||||
| NC score | 0.015136 (rank : 144) | θ value | 0.813845 (rank : 92) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00196, Q00197, Q00198, Q00199, Q00200, Q00201, Q05882, Q61995, Q61996, Q64245 | Gene names | Pou2f2, Oct2, Otf2 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.014751 (rank : 145) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
UTP20_HUMAN
|
||||||
| NC score | 0.014066 (rank : 146) | θ value | 2.36792 (rank : 113) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
YTHD3_HUMAN
|
||||||
| NC score | 0.014060 (rank : 147) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z739, Q63Z37, Q659A3 | Gene names | YTHDF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.013339 (rank : 148) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.011854 (rank : 149) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
IRX3_HUMAN
|
||||||
| NC score | 0.011692 (rank : 150) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78415, Q7Z4A4, Q7Z4A5, Q8IVC6 | Gene names | IRX3, IRXB1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
ABL2_HUMAN
|
||||||
| NC score | 0.011642 (rank : 151) | θ value | 0.0330416 (rank : 43) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
ROBO4_HUMAN
|
||||||
| NC score | 0.011498 (rank : 152) | θ value | 1.06291 (rank : 100) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WZ75, Q8TEG1, Q96JV6, Q9H718, Q9NWJ8 | Gene names | ROBO4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor (Magic roundabout). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.011097 (rank : 153) | θ value | 0.163984 (rank : 62) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
CRF_HUMAN
|
||||||
| NC score | 0.010984 (rank : 154) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06850 | Gene names | CRH | |||
|
Domain Architecture |

|
|||||
| Description | Corticoliberin precursor (Corticotropin-releasing factor) (CRF) (Corticotropin-releasing hormone). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.010913 (rank : 155) | θ value | 0.0961366 (rank : 56) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.010214 (rank : 156) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
AAK1_HUMAN
|
||||||
| NC score | 0.010021 (rank : 157) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.010008 (rank : 158) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.009795 (rank : 159) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.009744 (rank : 160) | θ value | 3.0926 (rank : 120) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.009698 (rank : 161) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.009695 (rank : 162) | θ value | 1.06291 (rank : 102) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
TMLH_HUMAN
|
||||||
| NC score | 0.008857 (rank : 163) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVH6 | Gene names | TMLHE, TMLH | |||
|
Domain Architecture |

|
|||||
| Description | Trimethyllysine dioxygenase, mitochondrial precursor (EC 1.14.11.8) (Epsilon-trimethyllysine 2-oxoglutarate dioxygenase) (TML-alpha- ketoglutarate dioxygenase) (TML hydroxylase) (TML dioxygenase) (TMLD). | |||||
|
WNK4_MOUSE
|
||||||
| NC score | 0.008482 (rank : 164) | θ value | 6.88961 (rank : 167) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.008210 (rank : 165) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
STX4_HUMAN
|
||||||
| NC score | 0.007992 (rank : 166) | θ value | 1.81305 (rank : 109) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12846, Q15525 | Gene names | STX4, STX4A | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-4 (NY-REN-31 antigen). | |||||
|
P66B_MOUSE
|
||||||
| NC score | 0.007659 (rank : 167) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHR5, Q8C9Q3 | Gene names | Gatad2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 beta (p66/p68) (GATA zinc finger domain- containing protein 2B). | |||||
|
GPR64_HUMAN
|
||||||
| NC score | 0.007608 (rank : 168) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.007230 (rank : 169) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
G3PT_HUMAN
|
||||||
| NC score | 0.006740 (rank : 170) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14556, O60823, Q6JTT9, Q9HCU6 | Gene names | GAPDHS, GAPD2, GAPDH2, GAPDS | |||
|
Domain Architecture |

|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
FGR_MOUSE
|
||||||
| NC score | 0.006493 (rank : 171) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14234, Q61404 | Gene names | Fgr | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FGR (EC 2.7.10.2) (P55-FGR) (C- FGR). | |||||
|
FGR_HUMAN
|
||||||
| NC score | 0.006406 (rank : 172) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P09769, Q9UIQ3 | Gene names | FGR, SRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FGR (EC 2.7.10.2) (P55-FGR) (C- FGR). | |||||
|
FYN_MOUSE
|
||||||
| NC score | 0.006293 (rank : 173) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39688 | Gene names | Fyn | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.006255 (rank : 174) | θ value | 0.279714 (rank : 70) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
TBC10_HUMAN
|
||||||
| NC score | 0.006207 (rank : 175) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXI6, O76053, Q543A2 | Gene names | TBC1D10A, EPI64, TBC1D10 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
FYN_HUMAN
|
||||||
| NC score | 0.005869 (rank : 176) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P06241 | Gene names | FYN | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn) (Protooncogene Syn) (SLK). | |||||
|
CCNB1_MOUSE
|
||||||
| NC score | 0.005842 (rank : 177) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
TSH3_MOUSE
|
||||||
| NC score | 0.005752 (rank : 178) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.005653 (rank : 179) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
KSR2_HUMAN
|
||||||
| NC score | 0.005482 (rank : 180) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6VAB6, Q8N775 | Gene names | KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-2 (hKSR2). | |||||
|
SSH1_HUMAN
|
||||||
| NC score | 0.005155 (rank : 181) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.004756 (rank : 182) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.004099 (rank : 183) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
M3K14_HUMAN
|
||||||
| NC score | 0.003573 (rank : 184) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99558, Q8IYN1 | Gene names | MAP3K14, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK) (HsNIK). | |||||
|
NRK_HUMAN
|
||||||
| NC score | 0.003573 (rank : 185) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.003250 (rank : 186) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.002678 (rank : 187) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
MTF1_HUMAN
|
||||||
| NC score | 0.000379 (rank : 188) | θ value | 0.47712 (rank : 82) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
ZXDA_HUMAN
|
||||||
| NC score | -0.001132 (rank : 189) | θ value | 1.38821 (rank : 105) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98168, Q9UJP7 | Gene names | ZXDA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDA. | |||||
|
KLF11_HUMAN
|
||||||
| NC score | -0.001221 (rank : 190) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14901 | Gene names | KLF11, FKLF, TIEG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 11 (Transforming growth factor-beta-inducible early growth response protein 2) (TGFB-inducible early growth response protein 2) (TIEG-2). | |||||
|
KLF11_MOUSE
|
||||||
| NC score | -0.001430 (rank : 191) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EPF4 | Gene names | Klf11, Tieg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 11 (Transforming growth factor-beta-inducible early growth response protein 2) (TGFB-inducible early growth response protein 2) (TIEG-2). | |||||