Please be patient as the page loads
|
TF65_HUMAN
|
||||||
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TF65_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TF65_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985470 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
REL_HUMAN
|
||||||
| θ value | 2.2976e-104 (rank : 3) | NC score | 0.934985 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
REL_MOUSE
|
||||||
| θ value | 8.17181e-102 (rank : 4) | NC score | 0.932120 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
RELB_MOUSE
|
||||||
| θ value | 4.81303e-86 (rank : 5) | NC score | 0.936205 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |
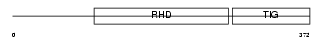
|
|||||
| Description | Transcription factor RelB. | |||||
|
RELB_HUMAN
|
||||||
| θ value | 6.28603e-86 (rank : 6) | NC score | 0.934719 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |
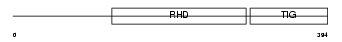
|
|||||
| Description | Transcription factor RelB (I-Rel). | |||||
|
NFKB2_HUMAN
|
||||||
| θ value | 1.6097e-65 (rank : 7) | NC score | 0.550478 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB2_MOUSE
|
||||||
| θ value | 2.10232e-65 (rank : 8) | NC score | 0.567887 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB1_MOUSE
|
||||||
| θ value | 6.32992e-62 (rank : 9) | NC score | 0.538509 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P25799, Q3TZE8, Q3V2V6, Q6TDG8, Q75ZL1, Q80Y21, Q8C712 | Gene names | Nfkb1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) (NF-kappa-B1 p84/NF-kappa-B1 p98) [Contains: Nuclear factor NF- kappa-B p50 subunit]. | |||||
|
NFKB1_HUMAN
|
||||||
| θ value | 6.99855e-61 (rank : 10) | NC score | 0.537644 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19838, Q68D84, Q86V43, Q8N4X7, Q9NZC0 | Gene names | NFKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) [Contains: Nuclear factor NF-kappa-B p50 subunit]. | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 11) | NC score | 0.265367 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 12) | NC score | 0.241006 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 13) | NC score | 0.261339 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 14) | NC score | 0.226233 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC2_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 15) | NC score | 0.210437 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 16) | NC score | 0.304976 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 17) | NC score | 0.311232 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.057962 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NKX23_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.023844 (rank : 154) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TAU0, Q9NYS6 | Gene names | NKX2-3, NKX23, NKX2C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.3 (Homeobox protein NKX-2.3) (Homeobox protein NK-2 homolog C). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.024200 (rank : 153) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.043395 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.052825 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.065819 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.029185 (rank : 136) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.036713 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.055351 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.027503 (rank : 141) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.056875 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.041918 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.066694 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NKX23_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.019145 (rank : 161) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97334, Q9QZ60, Q9WV67 | Gene names | Nkx2-3, Nkx-2.3, Nkx2c | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.3 (Homeobox protein NK-2 homolog C) (Nkx2-C) (Homeobox protein NK-2 homolog 3). | |||||
|
BUB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.022239 (rank : 158) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.035460 (rank : 126) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.024682 (rank : 148) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.031364 (rank : 133) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
IL7RA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.038978 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16872 | Gene names | Il7r | |||
|
Domain Architecture |
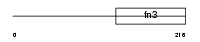
|
|||||
| Description | Interleukin-7 receptor alpha chain precursor (IL-7R-alpha) (CD127 antigen). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.024390 (rank : 151) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.047394 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.047376 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.006388 (rank : 184) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.043644 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.054286 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.036391 (rank : 122) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.029743 (rank : 135) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
3MG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.028774 (rank : 137) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29372, Q13770, Q15275, Q15961, Q96BZ6, Q96S33, Q9NNX5 | Gene names | MPG, AAG, ANPG, MID1 | |||
|
Domain Architecture |
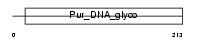
|
|||||
| Description | DNA-3-methyladenine glycosylase (EC 3.2.2.21) (3-methyladenine DNA glycosidase) (ADPG) (3-alkyladenine DNA glycosylase) (N-methylpurine- DNA glycosylase). | |||||
|
DTX1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.026598 (rank : 143) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |
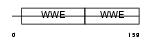
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.024638 (rank : 149) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.013773 (rank : 169) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.027247 (rank : 142) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.030617 (rank : 134) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.055547 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.061555 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
EVL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.027810 (rank : 140) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.037366 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
RAI2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.033318 (rank : 130) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
STA5A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.013161 (rank : 173) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.035590 (rank : 125) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.020989 (rank : 159) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.035405 (rank : 127) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.028622 (rank : 139) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
HDAC9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.014688 (rank : 168) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99N13, Q9EPT2 | Gene names | Hdac9, Hdac7b, Hdrp, Mitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (Histone deacetylase-related protein) (MEF2-interacting transcription repressor MITR). | |||||
|
NKX24_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.016837 (rank : 164) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H2Z4 | Gene names | NKX2-4, NKX2D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.4 (Homeobox protein NKX2.4) (Homeobox protein NK-2 homolog D). | |||||
|
NKX24_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.013679 (rank : 171) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQM3, Q9EQM4 | Gene names | Nkx2-4, Nkx2d | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.4 (Homeobox protein NKX2.4) (Homeobox protein NK-2 homolog D). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.026480 (rank : 144) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.047052 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PWWP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.026133 (rank : 145) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
RAI2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.041354 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.042563 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.019932 (rank : 160) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.036898 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.031680 (rank : 132) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.016476 (rank : 165) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
HD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.017586 (rank : 163) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
NRBP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.014957 (rank : 167) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V36, Q8R3M0 | Gene names | Nrbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding protein 2. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.033288 (rank : 131) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.033687 (rank : 129) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
DC1L2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.013358 (rank : 172) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.024290 (rank : 152) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.036417 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.008899 (rank : 178) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.055344 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN692_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | -0.000682 (rank : 192) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.022556 (rank : 157) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | -0.000159 (rank : 191) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.000128 (rank : 190) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.028623 (rank : 138) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
TBX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.005152 (rank : 187) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43435, O43436, Q96RJ2 | Gene names | TBX1 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.008658 (rank : 179) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.006949 (rank : 181) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.006470 (rank : 182) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
WASL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.036216 (rank : 123) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.041529 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.042482 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.023470 (rank : 155) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.018890 (rank : 162) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MPIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.007845 (rank : 180) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48964 | Gene names | Cdc25a, Cdc25m3 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 1 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25A). | |||||
|
MYPN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.004288 (rank : 188) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
PAPOB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.006345 (rank : 185) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRJ5, Q8NE14 | Gene names | PAPOLB, PAPT | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.006412 (rank : 183) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.057890 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.010134 (rank : 176) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.034306 (rank : 128) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
VGF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.025993 (rank : 147) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.002168 (rank : 189) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
AAKG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.010713 (rank : 175) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WG5, Q6V7V5 | Gene names | Prkag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.013679 (rank : 170) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.006145 (rank : 186) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CS016_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.040086 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
CT055_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.015441 (rank : 166) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R184, Q8BQE5, Q8K024, Q8VEM7, Q9CU80 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55 homolog. | |||||
|
FA61A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.009680 (rank : 177) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
FBLI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.010900 (rank : 174) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |
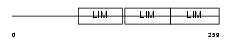
|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.024457 (rank : 150) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.023255 (rank : 156) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
K1683_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.036012 (rank : 124) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.026132 (rank : 146) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ABTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.050466 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ANFY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.054591 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANFY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.052950 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.053585 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.052899 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.058680 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.055940 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANKR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.053251 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANKR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.053931 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.063662 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ANKR7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.057412 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92527, Q96QN1, Q9UDM3 | Gene names | ANKRD7 | |||
|
Domain Architecture |
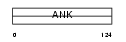
|
|||||
| Description | Ankyrin repeat domain-containing protein 7 (Testis-specific protein TSA806). | |||||
|
ANR10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.058806 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |
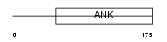
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.056862 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.057458 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
ANR23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.053881 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
ANR25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.055504 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
ANR28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052359 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.052652 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR33_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054742 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ANR41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.052140 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
ANR42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.066155 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR42_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.053286 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.052263 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANR52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.056121 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.057272 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANRA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.055076 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANRA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.054261 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ASB10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.050665 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.056207 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.050042 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
ASB16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.055351 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.065415 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.058084 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.058946 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.051782 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WWX0 | Gene names | ASB5 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 5 (ASB-5). | |||||
|
ASB6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.060699 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NWX5 | Gene names | ASB6 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ASB6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.067970 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |
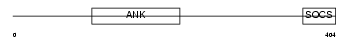
|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ASB7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.055172 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
Domain Architecture |
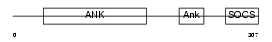
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.055069 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
Domain Architecture |
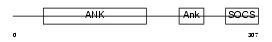
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.054007 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |
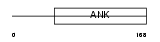
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ASB8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.053502 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ASB9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.054163 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DX5, Q9NWS5, Q9Y4T3 | Gene names | ASB9 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 9 (ASB-9). | |||||
|
BCL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.111609 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |
|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.112461 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CDN2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.051777 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.065480 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.066169 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.051813 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
IKBA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.124917 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25963 | Gene names | NFKBIA, IKBA, MAD3, NFKBI | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor alpha (Major histocompatibility complex enhancer- binding protein MAD3) (I-kappa-B-alpha) (IkappaBalpha) (IKB-alpha). | |||||
|
IKBA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.125684 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
IKBB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.126820 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
IKBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.124637 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
IKBE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.135791 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00221 | Gene names | NFKBIE, IKBE | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKBE). | |||||
|
IKBE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.129625 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
IKBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.071809 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
IKBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.070164 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88995, Q91X16, Q9R1Y3 | Gene names | Nfkbil1, Ikbl | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
INVS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.051778 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
INVS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.050707 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
MIB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.065440 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MIB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.065494 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
MIB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.059652 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
MIB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.056238 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.162989 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.192334 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
PA2G6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.067139 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PA2G6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.062958 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PSD10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.065545 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
PSD10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.065500 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
SNCAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.067046 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.059523 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.058786 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.051446 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.052535 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
ZDH13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.054562 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
ZDH13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.054148 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ZDH17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.057271 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ZDH17_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.056323 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |
|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
TF65_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TF65_MOUSE
|
||||||
| NC score | 0.985470 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
RELB_MOUSE
|
||||||
| NC score | 0.936205 (rank : 3) | θ value | 4.81303e-86 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB. | |||||
|
REL_HUMAN
|
||||||
| NC score | 0.934985 (rank : 4) | θ value | 2.2976e-104 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
RELB_HUMAN
|
||||||
| NC score | 0.934719 (rank : 5) | θ value | 6.28603e-86 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB (I-Rel). | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.932120 (rank : 6) | θ value | 8.17181e-102 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
NFKB2_MOUSE
|
||||||
| NC score | 0.567887 (rank : 7) | θ value | 2.10232e-65 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB2_HUMAN
|
||||||
| NC score | 0.550478 (rank : 8) | θ value | 1.6097e-65 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB1_MOUSE
|
||||||
| NC score | 0.538509 (rank : 9) | θ value | 6.32992e-62 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P25799, Q3TZE8, Q3V2V6, Q6TDG8, Q75ZL1, Q80Y21, Q8C712 | Gene names | Nfkb1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) (NF-kappa-B1 p84/NF-kappa-B1 p98) [Contains: Nuclear factor NF- kappa-B p50 subunit]. | |||||
|
NFKB1_HUMAN
|
||||||
| NC score | 0.537644 (rank : 10) | θ value | 6.99855e-61 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19838, Q68D84, Q86V43, Q8N4X7, Q9NZC0 | Gene names | NFKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) [Contains: Nuclear factor NF-kappa-B p50 subunit]. | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.311232 (rank : 11) | θ value | 2.44474e-05 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.304976 (rank : 12) | θ value | 2.44474e-05 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.265367 (rank : 13) | θ value | 3.64472e-09 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.261339 (rank : 14) | θ value | 5.26297e-08 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.241006 (rank : 15) | θ value | 1.80886e-08 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC1_MOUSE
|
||||||
| NC score | 0.226233 (rank : 16) | θ value | 3.77169e-06 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC2_MOUSE
|
||||||
| NC score | 0.210437 (rank : 17) | θ value | 1.09739e-05 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.192334 (rank : 18) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.162989 (rank : 19) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
IKBE_HUMAN
|
||||||
| NC score | 0.135791 (rank : 20) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00221 | Gene names | NFKBIE, IKBE | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKBE). | |||||
|
IKBE_MOUSE
|
||||||
| NC score | 0.129625 (rank : 21) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
IKBB_HUMAN
|
||||||
| NC score | 0.126820 (rank : 22) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
IKBA_MOUSE
|
||||||
| NC score | 0.125684 (rank : 23) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
IKBA_HUMAN
|
||||||
| NC score | 0.124917 (rank : 24) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25963 | Gene names | NFKBIA, IKBA, MAD3, NFKBI | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor alpha (Major histocompatibility complex enhancer- binding protein MAD3) (I-kappa-B-alpha) (IkappaBalpha) (IKB-alpha). | |||||
|
IKBB_MOUSE
|
||||||
| NC score | 0.124637 (rank : 25) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.112461 (rank : 26) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
BCL3_HUMAN
|
||||||
| NC score | 0.111609 (rank : 27) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
IKBL_HUMAN
|
||||||
| NC score | 0.071809 (rank : 28) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
IKBL_MOUSE
|
||||||
| NC score | 0.070164 (rank : 29) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88995, Q91X16, Q9R1Y3 | Gene names | Nfkbil1, Ikbl | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
ASB6_MOUSE
|
||||||
| NC score | 0.067970 (rank : 30) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
PA2G6_HUMAN
|
||||||
| NC score | 0.067139 (rank : 31) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
SNCAP_HUMAN
|
||||||
| NC score | 0.067046 (rank : 32) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.066694 (rank : 33) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.066169 (rank : 34) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ANR42_HUMAN
|
||||||
| NC score | 0.066155 (rank : 35) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.065819 (rank : 36) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PSD10_HUMAN
|
||||||
| NC score | 0.065545 (rank : 37) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
PSD10_MOUSE
|
||||||
| NC score | 0.065500 (rank : 38) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
MIB1_MOUSE
|
||||||
| NC score | 0.065494 (rank : 39) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.065480 (rank : 40) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MIB1_HUMAN
|
||||||
| NC score | 0.065440 (rank : 41) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
ASB16_MOUSE
|
||||||
| NC score | 0.065415 (rank : 42) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.063662 (rank : 43) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
PA2G6_MOUSE
|
||||||
| NC score | 0.062958 (rank : 44) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.061555 (rank : 45) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
ASB6_HUMAN
|
||||||
| NC score | 0.060699 (rank : 46) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NWX5 | Gene names | ASB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.059652 (rank : 47) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.059523 (rank : 48) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ASB3_MOUSE
|
||||||
| NC score | 0.058946 (rank : 49) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ANR10_HUMAN
|
||||||
| NC score | 0.058806 (rank : 50) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.058786 (rank : 51) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.058680 (rank : 52) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ASB3_HUMAN
|
||||||
| NC score | 0.058084 (rank : 53) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.057962 (rank : 54) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.057890 (rank : 55) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
ANR23_HUMAN
|
||||||
| NC score | 0.057458 (rank : 56) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
ANKR7_HUMAN
|
||||||
| NC score | 0.057412 (rank : 57) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92527, Q96QN1, Q9UDM3 | Gene names | ANKRD7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 7 (Testis-specific protein TSA806). | |||||
|
ANR52_MOUSE
|
||||||
| NC score | 0.057272 (rank : 58) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ZDH17_HUMAN
|
||||||
| NC score | 0.057271 (rank : 59) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.056875 (rank : 60) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ANR10_MOUSE
|
||||||
| NC score | 0.056862 (rank : 61) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ZDH17_MOUSE
|
||||||
| NC score | 0.056323 (rank : 62) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
MIB2_MOUSE
|
||||||
| NC score | 0.056238 (rank : 63) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
ASB10_MOUSE
|
||||||
| NC score | 0.056207 (rank : 64) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ANR52_HUMAN
|
||||||
| NC score | 0.056121 (rank : 65) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.055940 (rank : 66) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.055547 (rank : 67) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ANR25_MOUSE
|
||||||
| NC score | 0.055504 (rank : 68) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.055351 (rank : 69) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
ASB16_HUMAN
|
||||||
| NC score | 0.055351 (rank : 70) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.055344 (rank : 71) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ASB7_HUMAN
|
||||||
| NC score | 0.055172 (rank : 72) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ANRA2_HUMAN
|
||||||
| NC score | 0.055076 (rank : 73) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ASB7_MOUSE
|
||||||
| NC score | 0.055069 (rank : 74) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ANR33_HUMAN
|
||||||
| NC score | 0.054742 (rank : 75) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ANFY1_HUMAN
|
||||||
| NC score | 0.054591 (rank : 76) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ZDH13_HUMAN
|
||||||
| NC score | 0.054562 (rank : 77) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.054286 (rank : 78) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ANRA2_MOUSE
|
||||||
| NC score | 0.054261 (rank : 79) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ASB9_HUMAN
|
||||||
| NC score | 0.054163 (rank : 80) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DX5, Q9NWS5, Q9Y4T3 | Gene names | ASB9 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 9 (ASB-9). | |||||
|
ZDH13_MOUSE
|
||||||
| NC score | 0.054148 (rank : 81) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ASB8_HUMAN
|
||||||
| NC score | 0.054007 (rank : 82) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ANKR1_MOUSE
|
||||||
| NC score | 0.053931 (rank : 83) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANR23_MOUSE
|
||||||
| NC score | 0.053881 (rank : 84) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.053585 (rank : 85) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ASB8_MOUSE
|
||||||
| NC score | 0.053502 (rank : 86) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ANR42_MOUSE
|
||||||
| NC score | 0.053286 (rank : 87) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANKR1_HUMAN
|
||||||
| NC score | 0.053251 (rank : 88) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANFY1_MOUSE
|
||||||
| NC score | 0.052950 (rank : 89) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.052899 (rank : 90) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.052825 (rank : 91) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.052652 (rank : 92) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.052535 (rank : 93) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
ANR28_HUMAN
|
||||||
| NC score | 0.052359 (rank : 94) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR44_HUMAN
|
||||||
| NC score | 0.052263 (rank : 95) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANR41_HUMAN
|
||||||
| NC score | 0.052140 (rank : 96) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.051813 (rank : 97) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
ASB5_HUMAN
|
||||||
| NC score | 0.051782 (rank : 98) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WWX0 | Gene names | ASB5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 5 (ASB-5). | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.051778 (rank : 99) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
CDN2D_MOUSE
|
||||||
| NC score | 0.051777 (rank : 100) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.051446 (rank : 101) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.050707 (rank : 102) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
ASB10_HUMAN
|
||||||
| NC score | 0.050665 (rank : 103) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ABTB2_HUMAN
|
||||||
| NC score | 0.050466 (rank : 104) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ASB14_MOUSE
|
||||||
| NC score | 0.050042 (rank : 105) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.047394 (rank : 106) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.047376 (rank : 107) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.047052 (rank : 108) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.043644 (rank : 109) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.043395 (rank : 110) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.042563 (rank : 111) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.042482 (rank : 112) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.041918 (rank : 113) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.041529 (rank : 114) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
RAI2_MOUSE
|
||||||
| NC score | 0.041354 (rank : 115) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.040086 (rank : 116) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
IL7RA_MOUSE
|
||||||
| NC score | 0.038978 (rank : 117) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16872 | Gene names | Il7r | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-7 receptor alpha chain precursor (IL-7R-alpha) (CD127 antigen). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.037366 (rank : 118) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.036898 (rank : 119) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.036713 (rank : 120) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.036417 (rank : 121) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.036391 (rank : 122) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.036216 (rank : 123) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.036012 (rank : 124) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.035590 (rank : 125) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.035460 (rank : 126) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.035405 (rank : 127) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.034306 (rank : 128) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.033687 (rank : 129) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
RAI2_HUMAN
|
||||||
| NC score | 0.033318 (rank : 130) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.033288 (rank : 131) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.031680 (rank : 132) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.031364 (rank : 133) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.030617 (rank : 134) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.029743 (rank : 135) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.029185 (rank : 136) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
3MG_HUMAN
|
||||||
| NC score | 0.028774 (rank : 137) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29372, Q13770, Q15275, Q15961, Q96BZ6, Q96S33, Q9NNX5 | Gene names | MPG, AAG, ANPG, MID1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-3-methyladenine glycosylase (EC 3.2.2.21) (3-methyladenine DNA glycosidase) (ADPG) (3-alkyladenine DNA glycosylase) (N-methylpurine- DNA glycosylase). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.028623 (rank : 138) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.028622 (rank : 139) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.027810 (rank : 140) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.027503 (rank : 141) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.027247 (rank : 142) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
DTX1_MOUSE
|
||||||
| NC score | 0.026598 (rank : 143) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.026480 (rank : 144) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
PWWP2_HUMAN
|
||||||
| NC score | 0.026133 (rank : 145) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.026132 (rank : 146) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
VGF_HUMAN
|
||||||
| NC score | 0.025993 (rank : 147) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.024682 (rank : 148) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.024638 (rank : 149) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.024457 (rank : 150) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.024390 (rank : 151) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.024290 (rank : 152) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.024200 (rank : 153) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NKX23_HUMAN
|
||||||
| NC score | 0.023844 (rank : 154) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TAU0, Q9NYS6 | Gene names | NKX2-3, NKX23, NKX2C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.3 (Homeobox protein NKX-2.3) (Homeobox protein NK-2 homolog C). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.023470 (rank : 155) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.023255 (rank : 156) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.022556 (rank : 157) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
BUB1_HUMAN
|
||||||
| NC score | 0.022239 (rank : 158) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.020989 (rank : 159) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.019932 (rank : 160) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
NKX23_MOUSE
|
||||||
| NC score | 0.019145 (rank : 161) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97334, Q9QZ60, Q9WV67 | Gene names | Nkx2-3, Nkx-2.3, Nkx2c | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.3 (Homeobox protein NK-2 homolog C) (Nkx2-C) (Homeobox protein NK-2 homolog 3). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.018890 (rank : 162) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.017586 (rank : 163) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
NKX24_HUMAN
|
||||||
| NC score | 0.016837 (rank : 164) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H2Z4 | Gene names | NKX2-4, NKX2D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.4 (Homeobox protein NKX2.4) (Homeobox protein NK-2 homolog D). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.016476 (rank : 165) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
CT055_MOUSE
|
||||||
| NC score | 0.015441 (rank : 166) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R184, Q8BQE5, Q8K024, Q8VEM7, Q9CU80 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55 homolog. | |||||
|
NRBP2_MOUSE
|
||||||
| NC score | 0.014957 (rank : 167) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V36, Q8R3M0 | Gene names | Nrbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding protein 2. | |||||
|
HDAC9_MOUSE
|
||||||
| NC score | 0.014688 (rank : 168) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99N13, Q9EPT2 | Gene names | Hdac9, Hdac7b, Hdrp, Mitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (Histone deacetylase-related protein) (MEF2-interacting transcription repressor MITR). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.013773 (rank : 169) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.013679 (rank : 170) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
NKX24_MOUSE
|
||||||
| NC score | 0.013679 (rank : 171) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQM3, Q9EQM4 | Gene names | Nkx2-4, Nkx2d | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.4 (Homeobox protein NKX2.4) (Homeobox protein NK-2 homolog D). | |||||
|
DC1L2_MOUSE
|
||||||
| NC score | 0.013358 (rank : 172) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
STA5A_MOUSE
|
||||||
| NC score | 0.013161 (rank : 173) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
FBLI1_HUMAN
|
||||||
| NC score | 0.010900 (rank : 174) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
AAKG2_MOUSE
|
||||||
| NC score | 0.010713 (rank : 175) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WG5, Q6V7V5 | Gene names | Prkag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2). | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.010134 (rank : 176) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
FA61A_MOUSE
|
||||||
| NC score | 0.009680 (rank : 177) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.008899 (rank : 178) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.008658 (rank : 179) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
MPIP1_MOUSE
|
||||||
| NC score | 0.007845 (rank : 180) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48964 | Gene names | Cdc25a, Cdc25m3 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 1 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25A). | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.006949 (rank : 181) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.006470 (rank : 182) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.006412 (rank : 183) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.006388 (rank : 184) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PAPOB_HUMAN
|
||||||
| NC score | 0.006345 (rank : 185) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRJ5, Q8NE14 | Gene names | PAPOLB, PAPT | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.006145 (rank : 186) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
TBX1_HUMAN
|
||||||
| NC score | 0.005152 (rank : 187) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43435, O43436, Q96RJ2 | Gene names | TBX1 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
MYPN_HUMAN
|
||||||
| NC score | 0.004288 (rank : 188) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.002168 (rank : 189) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.000128 (rank : 190) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | -0.000159 (rank : 191) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
ZN692_MOUSE
|
||||||
| NC score | -0.000682 (rank : 192) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||