Please be patient as the page loads
|
CXXC6_HUMAN
|
||||||
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CXXC6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 180 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
DNMT1_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 2) | NC score | 0.252909 (rank : 2) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 3) | NC score | 0.173063 (rank : 8) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 4) | NC score | 0.240539 (rank : 3) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
WNK4_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 5) | NC score | 0.027553 (rank : 116) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 6) | NC score | 0.120606 (rank : 14) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
CXCC1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 7) | NC score | 0.199200 (rank : 6) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |
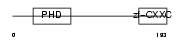
|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
MBD1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.214275 (rank : 4) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UIS9, O15248, O95241, Q7Z7B5, Q8N4W4, Q9UNZ6, Q9UNZ7, Q9UNZ8, Q9UNZ9 | Gene names | MBD1, PCM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1) (Protein containing methyl-CpG-binding domain 1). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 9) | NC score | 0.088418 (rank : 18) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
MBD1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 10) | NC score | 0.209014 (rank : 5) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z2E2, Q792D6, Q8CCL9, Q9DC19 | Gene names | Mbd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 11) | NC score | 0.111878 (rank : 15) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.082630 (rank : 19) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.023763 (rank : 130) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
CXCC1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.192502 (rank : 7) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
JHD1B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.130860 (rank : 13) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NHM5, Q8NCI2, Q96HC7, Q96SL0, Q96T03, Q9NS96, Q9UF75 | Gene names | FBXL10, CXXC2, FBL10, JHDM1B, PCCX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10) (Protein JEMMA) (Jumonji domain-containing EMSY-interactor methyltransferase motif protein) (CXXC-type zinc finger protein 2) (Protein-containing CXXC domain 2). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.132152 (rank : 12) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.145382 (rank : 9) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.061525 (rank : 46) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
PRIC2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.069659 (rank : 31) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z3G6 | Gene names | PRICKLE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
PRIC2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.069829 (rank : 30) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80Y24 | Gene names | Prickle2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.031606 (rank : 105) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.044894 (rank : 81) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.071841 (rank : 28) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NBEA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.035240 (rank : 99) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
NBN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.066289 (rank : 38) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.055841 (rank : 57) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
MAGAA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.023793 (rank : 129) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |
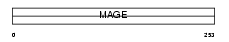
|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.063170 (rank : 40) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.038533 (rank : 94) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
CAF1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.027412 (rank : 117) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D0N7 | Gene names | Chaf1b | |||
|
Domain Architecture |
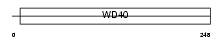
|
|||||
| Description | Chromatin assembly factor 1 subunit B (CAF-1 subunit B) (Chromatin assembly factor I p60 subunit) (CAF-I 60 kDa subunit) (CAF-Ip60). | |||||
|
FRMD5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.022001 (rank : 134) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.060232 (rank : 50) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.066574 (rank : 37) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PRIC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.061787 (rank : 44) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96MT3, Q71QF8, Q96N00 | Gene names | PRICKLE1, RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 1 (REST/NRSF-interacting LIM domain protein 1). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.082401 (rank : 20) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.061253 (rank : 47) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
BMP3B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.023987 (rank : 128) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97737 | Gene names | Gdf10, Bmp3b | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.029797 (rank : 111) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HSF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.053763 (rank : 62) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.062443 (rank : 42) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.016151 (rank : 158) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.038703 (rank : 93) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.069603 (rank : 32) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.055423 (rank : 60) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.059413 (rank : 53) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
IPYR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.045743 (rank : 77) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15181, Q5SQT7, Q6P7P4, Q9UQJ5, Q9Y5B1 | Gene names | PPA1, IOPPP, PP | |||
|
Domain Architecture |

|
|||||
| Description | Inorganic pyrophosphatase (EC 3.6.1.1) (Pyrophosphate phospho- hydrolase) (PPase). | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.030569 (rank : 107) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.045028 (rank : 80) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.072575 (rank : 26) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.072363 (rank : 27) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.075357 (rank : 24) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
EGR3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.009672 (rank : 179) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43300, Q9R276 | Gene names | Egr3, Egr-3 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 3 (EGR-3). | |||||
|
EPM2A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.045729 (rank : 78) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
FXL19_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.134211 (rank : 11) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PCT2, Q8N789, Q9NT14 | Gene names | FBXL19, FBL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
FXL19_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.134593 (rank : 10) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.053173 (rank : 64) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.022899 (rank : 132) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.022371 (rank : 133) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
FOXJ3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.015726 (rank : 162) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |
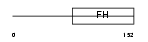
|
|||||
| Description | Forkhead box protein J3. | |||||
|
GP1BA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.021805 (rank : 135) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
K0513_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.060010 (rank : 51) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60268, Q8N6G0 | Gene names | KIAA0513 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0513. | |||||
|
K0513_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.060562 (rank : 49) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0A7, Q80TY8, Q8BQV3, Q8C8N9, Q8CCN0 | Gene names | Kiaa0513 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0513. | |||||
|
K1C18_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.009928 (rank : 178) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05783, Q9BW26 | Gene names | KRT18, CYK18 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 18 (Cytokeratin-18) (CK-18) (Keratin-18) (K18). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.042307 (rank : 85) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NUMB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.032783 (rank : 103) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
PLCB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.024567 (rank : 127) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.044809 (rank : 82) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
SON_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.076054 (rank : 23) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.060905 (rank : 48) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TERF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.048045 (rank : 76) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |
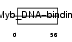
|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TMM24_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.037166 (rank : 96) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.049348 (rank : 74) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.029515 (rank : 112) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.024762 (rank : 126) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
DPOLZ_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.034964 (rank : 100) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
EGR3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.008837 (rank : 183) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06889 | Gene names | EGR3, PILOT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 3 (EGR-3) (Zinc finger protein pilot). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.040761 (rank : 88) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.042129 (rank : 86) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.041783 (rank : 87) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.023436 (rank : 131) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.043174 (rank : 84) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SEM6D_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.014853 (rank : 164) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEMG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.039968 (rank : 89) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.079865 (rank : 21) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TF65_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.028622 (rank : 113) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.017022 (rank : 156) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TR10C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.030047 (rank : 109) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |
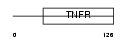
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
USH2A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.012332 (rank : 174) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.012960 (rank : 173) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.036851 (rank : 97) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
ZN185_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.051578 (rank : 68) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
APLP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.026011 (rank : 121) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
CHSTC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.020999 (rank : 140) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRB3, Q9NXY7 | Gene names | CHST12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2) (Sulfotransferase Hlo). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.057737 (rank : 55) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.032276 (rank : 104) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JHD1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.110447 (rank : 17) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2K7, Q49A21, Q4G0M3, Q69YY8, Q9BVH5, Q9H7H5, Q9UK66 | Gene names | FBXL11, FBL7, JHDM1A, KIAA1004 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11) (F-box protein FBL7) (F-box protein Lilina). | |||||
|
JHD1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.111073 (rank : 16) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59997, Q3U1M5, Q3UR56, Q3V3Q1, Q69ZT4 | Gene names | Fbxl11, Jhdm1a, Kiaa1004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.050503 (rank : 71) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NUPL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.036511 (rank : 98) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
PIWL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.017175 (rank : 153) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TC59, Q96SW6, Q9NW28 | Gene names | PIWIL2, HILI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.014689 (rank : 166) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.015441 (rank : 163) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
ZN286_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.002477 (rank : 196) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HBT8, Q96JF3 | Gene names | ZNF286, KIAA1874 | |||
|
Domain Architecture |
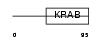
|
|||||
| Description | Zinc finger protein 286. | |||||
|
ZN294_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.028522 (rank : 115) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
ABI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.021481 (rank : 137) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.021173 (rank : 139) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
CENG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.009145 (rank : 181) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
GA2L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.026857 (rank : 118) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
GUC2C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.004598 (rank : 193) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.067672 (rank : 35) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MTG8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.030547 (rank : 108) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
MTG8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.030581 (rank : 106) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.044415 (rank : 83) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
PHC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.028533 (rank : 114) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.069948 (rank : 29) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.037417 (rank : 95) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.008255 (rank : 186) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
SOX4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.015828 (rank : 161) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.069084 (rank : 33) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.014766 (rank : 165) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.039944 (rank : 90) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
ZCH14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.034418 (rank : 101) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ZN509_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.003496 (rank : 195) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.026585 (rank : 119) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CNTFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.025621 (rank : 122) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26992 | Gene names | CNTFR | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary neurotrophic factor receptor alpha precursor (CNTFR alpha). | |||||
|
CNTFR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.025434 (rank : 123) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88507, Q80T01 | Gene names | Cntfr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary neurotrophic factor receptor alpha precursor (CNTFR alpha). | |||||
|
ESR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.017230 (rank : 151) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P03372, Q13511, Q14276, Q9NU51, Q9UDZ7, Q9UIS7 | Gene names | ESR1, ESR, NR3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
ESR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.017177 (rank : 152) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19785, Q9JJT5, Q9QY51, Q9QY52 | Gene names | Esr1, Esr, Estr, Estra, Nr3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
ESR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.017114 (rank : 155) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92731, O60608, O60685, O60702, O60703, O75583, O75584, Q86Z31, Q9UEV6, Q9UHD3, Q9UQK9 | Gene names | ESR2, ESTRB, NR3A2 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor beta (ER-beta). | |||||
|
ESR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.017260 (rank : 150) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08537, O35635, O70519 | Gene names | Esr2, Estrb, Nr3a2 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor beta (ER-beta). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.018240 (rank : 147) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.008637 (rank : 184) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.045510 (rank : 79) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.019638 (rank : 143) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PPP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.009134 (rank : 182) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.039308 (rank : 91) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
RGS11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.014183 (rank : 169) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |
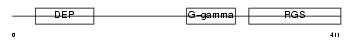
|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.075202 (rank : 25) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.062663 (rank : 41) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.069004 (rank : 34) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.057563 (rank : 56) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.020564 (rank : 142) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
ZN682_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.001718 (rank : 197) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95780, Q96JV9 | Gene names | ZNF682 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 682. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.038756 (rank : 92) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.032795 (rank : 102) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
CECR6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.026069 (rank : 120) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXQ6 | Gene names | CECR6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
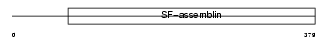
| Description | Cat eye syndrome critical region protein 6. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.007294 (rank : 188) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
HOMEZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.019416 (rank : 144) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
JADE3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.021270 (rank : 138) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
K1C18_MOUSE
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.007439 (rank : 187) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05784, Q61766 | Gene names | Krt18, Kerd, Krt1-18 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 18 (Cytokeratin-18) (CK-18) (Keratin-18) (K18) (Cytokeratin endo B) (Keratin D). | |||||
|
LRC8E_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.005716 (rank : 189) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NSJ5, Q2YDY3, Q7L236, Q8N3B0, Q9H5H8 | Gene names | LRRC8E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8E. | |||||
|
NGN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.012140 (rank : 175) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.014051 (rank : 170) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.008469 (rank : 185) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
AMZ1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.015865 (rank : 160) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q400G9, Q8TF51 | Gene names | AMZ1, KIAA1950 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Archaemetzincin-1 (EC 3.-.-.-) (Archeobacterial metalloproteinase-like protein 1). | |||||
|
BAI2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.005384 (rank : 191) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.004274 (rank : 194) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.048316 (rank : 75) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CU013_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.015975 (rank : 159) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.021754 (rank : 136) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DMBT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.004958 (rank : 192) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.009161 (rank : 180) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.013466 (rank : 171) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
IPYR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.029809 (rank : 110) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D819 | Gene names | Ppa1, Pp, Pyp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase (EC 3.6.1.1) (Pyrophosphate phospho- hydrolase) (PPase). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.018131 (rank : 148) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
LCP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.020747 (rank : 141) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60787 | Gene names | Lcp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.014299 (rank : 168) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
LYRIC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.019398 (rank : 145) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
NOP56_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.019342 (rank : 146) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
OTX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.005500 (rank : 190) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80205 | Gene names | Otx1, Otx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.052325 (rank : 66) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
PPARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.010152 (rank : 176) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07869, Q16241, Q92486, Q92689, Q9Y3N1 | Gene names | PPARA, NR1C1, PPAR | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor alpha (PPAR-alpha). | |||||
|
PROM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.016653 (rank : 157) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54990, O35408 | Gene names | Prom1, Prom, Proml1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133 homolog). | |||||
|
PXR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.010017 (rank : 177) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75469, Q96AC7, Q9UJ22, Q9UJ23, Q9UJ24, Q9UJ25, Q9UJ26, Q9UJ27 | Gene names | NR1I2, PXR | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor PXR (Pregnane X receptor) (Orphan nuclear receptor PAR1) (Steroid and xenobiotic receptor) (SXR). | |||||
|
RMI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.017122 (rank : 154) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9A7, Q5SQG8, Q5SQG9, Q6P1Q4, Q6PI89, Q7Z6L6 | Gene names | RMI1, C9orf76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RMI1 homolog. | |||||
|
SCAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.014604 (rank : 167) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
SKIL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.013449 (rank : 172) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
UBAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.017425 (rank : 149) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ09, Q4V759, Q5T7B3, Q6FI75, Q8NC52, Q8NCG6, Q8NCH9 | Gene names | UBAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 1 (UBAP). | |||||
|
UBL7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.025270 (rank : 125) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96S82, Q96I03 | Gene names | UBL7, BMSCUBP | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-like protein 7 (Ubiquitin-like protein SB132) (Bone marrow stromal cell ubiquitin-like protein) (BMSC-UbP). | |||||
|
UBL7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.025412 (rank : 124) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.050093 (rank : 73) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
K1718_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.053214 (rank : 63) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
K1718_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.055629 (rank : 59) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.051044 (rank : 70) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.077561 (rank : 22) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.059320 (rank : 54) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.062045 (rank : 43) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.051261 (rank : 69) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.067376 (rank : 36) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NFH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.050311 (rank : 72) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PHF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.061604 (rank : 45) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
PHF8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.055029 (rank : 61) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
PHF8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.053123 (rank : 65) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80TJ7, Q8BLX8, Q8BLY0, Q8BZ61, Q8CG26 | Gene names | Phf8, Kiaa1111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.059838 (rank : 52) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.055669 (rank : 58) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.064274 (rank : 39) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.051681 (rank : 67) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 180 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
DNMT1_MOUSE
|
||||||
| NC score | 0.252909 (rank : 2) | θ value | 4.1701e-05 (rank : 2) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.240539 (rank : 3) | θ value | 5.44631e-05 (rank : 4) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
MBD1_HUMAN
|
||||||
| NC score | 0.214275 (rank : 4) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UIS9, O15248, O95241, Q7Z7B5, Q8N4W4, Q9UNZ6, Q9UNZ7, Q9UNZ8, Q9UNZ9 | Gene names | MBD1, PCM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1) (Protein containing methyl-CpG-binding domain 1). | |||||
|
MBD1_MOUSE
|
||||||
| NC score | 0.209014 (rank : 5) | θ value | 0.00869519 (rank : 10) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z2E2, Q792D6, Q8CCL9, Q9DC19 | Gene names | Mbd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1). | |||||
|
CXCC1_HUMAN
|
||||||
| NC score | 0.199200 (rank : 6) | θ value | 0.00509761 (rank : 7) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
CXCC1_MOUSE
|
||||||
| NC score | 0.192502 (rank : 7) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.173063 (rank : 8) | θ value | 4.1701e-05 (rank : 3) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.145382 (rank : 9) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
FXL19_MOUSE
|
||||||
| NC score | 0.134593 (rank : 10) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
FXL19_HUMAN
|
||||||
| NC score | 0.134211 (rank : 11) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PCT2, Q8N789, Q9NT14 | Gene names | FBXL19, FBL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.132152 (rank : 12) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
JHD1B_HUMAN
|
||||||
| NC score | 0.130860 (rank : 13) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NHM5, Q8NCI2, Q96HC7, Q96SL0, Q96T03, Q9NS96, Q9UF75 | Gene names | FBXL10, CXXC2, FBL10, JHDM1B, PCCX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10) (Protein JEMMA) (Jumonji domain-containing EMSY-interactor methyltransferase motif protein) (CXXC-type zinc finger protein 2) (Protein-containing CXXC domain 2). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.120606 (rank : 14) | θ value | 0.00390308 (rank : 6) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.111878 (rank : 15) | θ value | 0.00869519 (rank : 11) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
JHD1A_MOUSE
|
||||||
| NC score | 0.111073 (rank : 16) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59997, Q3U1M5, Q3UR56, Q3V3Q1, Q69ZT4 | Gene names | Fbxl11, Jhdm1a, Kiaa1004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11). | |||||
|
JHD1A_HUMAN
|
||||||
| NC score | 0.110447 (rank : 17) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2K7, Q49A21, Q4G0M3, Q69YY8, Q9BVH5, Q9H7H5, Q9UK66 | Gene names | FBXL11, FBL7, JHDM1A, KIAA1004 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11) (F-box protein FBL7) (F-box protein Lilina). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.088418 (rank : 18) | θ value | 0.00665767 (rank : 9) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.082630 (rank : 19) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.082401 (rank : 20) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.079865 (rank : 21) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.077561 (rank : 22) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.076054 (rank : 23) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.075357 (rank : 24) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.075202 (rank : 25) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.072575 (rank : 26) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.072363 (rank : 27) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.071841 (rank : 28) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.069948 (rank : 29) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
PRIC2_MOUSE
|
||||||
| NC score | 0.069829 (rank : 30) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80Y24 | Gene names | Prickle2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
PRIC2_HUMAN
|
||||||
| NC score | 0.069659 (rank : 31) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z3G6 | Gene names | PRICKLE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.069603 (rank : 32) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.069084 (rank : 33) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.069004 (rank : 34) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.067672 (rank : 35) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.067376 (rank : 36) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.066574 (rank : 37) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NBN_HUMAN
|
||||||
| NC score | 0.066289 (rank : 38) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.064274 (rank : 39) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.063170 (rank : 40) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.062663 (rank : 41) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.062443 (rank : 42) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.062045 (rank : 43) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PRIC1_HUMAN
|
||||||
| NC score | 0.061787 (rank : 44) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96MT3, Q71QF8, Q96N00 | Gene names | PRICKLE1, RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 1 (REST/NRSF-interacting LIM domain protein 1). | |||||
|
PHF2_HUMAN
|
||||||
| NC score | 0.061604 (rank : 45) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.061525 (rank : 46) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.061253 (rank : 47) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.060905 (rank : 48) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
K0513_MOUSE
|
||||||
| NC score | 0.060562 (rank : 49) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0A7, Q80TY8, Q8BQV3, Q8C8N9, Q8CCN0 | Gene names | Kiaa0513 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0513. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.060232 (rank : 50) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
K0513_HUMAN
|
||||||
| NC score | 0.060010 (rank : 51) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60268, Q8N6G0 | Gene names | KIAA0513 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0513. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.059838 (rank : 52) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.059413 (rank : 53) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.059320 (rank : 54) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.057737 (rank : 55) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.057563 (rank : 56) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.055841 (rank : 57) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.055669 (rank : 58) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
K1718_MOUSE
|
||||||
| NC score | 0.055629 (rank : 59) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.055423 (rank : 60) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PHF8_HUMAN
|
||||||
| NC score | 0.055029 (rank : 61) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
HSF1_HUMAN
|
||||||
| NC score | 0.053763 (rank : 62) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
K1718_HUMAN
|
||||||
| NC score | 0.053214 (rank : 63) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.053173 (rank : 64) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
PHF8_MOUSE
|
||||||
| NC score | 0.053123 (rank : 65) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80TJ7, Q8BLX8, Q8BLY0, Q8BZ61, Q8CG26 | Gene names | Phf8, Kiaa1111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.052325 (rank : 66) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.051681 (rank : 67) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZN185_HUMAN
|
||||||
| NC score | 0.051578 (rank : 68) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.051261 (rank : 69) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.051044 (rank : 70) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.050503 (rank : 71) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.050311 (rank : 72) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.050093 (rank : 73) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.049348 (rank : 74) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.048316 (rank : 75) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TERF2_HUMAN
|
||||||
| NC score | 0.048045 (rank : 76) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
IPYR_HUMAN
|
||||||
| NC score | 0.045743 (rank : 77) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15181, Q5SQT7, Q6P7P4, Q9UQJ5, Q9Y5B1 | Gene names | PPA1, IOPPP, PP | |||
|
Domain Architecture |

|
|||||
| Description | Inorganic pyrophosphatase (EC 3.6.1.1) (Pyrophosphate phospho- hydrolase) (PPase). | |||||
|
EPM2A_MOUSE
|
||||||
| NC score | 0.045729 (rank : 78) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.045510 (rank : 79) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.045028 (rank : 80) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.044894 (rank : 81) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.044809 (rank : 82) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.044415 (rank : 83) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.043174 (rank : 84) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.042307 (rank : 85) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.042129 (rank : 86) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.041783 (rank : 87) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.040761 (rank : 88) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
SEMG1_HUMAN
|
||||||
| NC score | 0.039968 (rank : 89) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.039944 (rank : 90) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.039308 (rank : 91) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.038756 (rank : 92) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.038703 (rank : 93) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.038533 (rank : 94) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.037417 (rank : 95) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TMM24_MOUSE
|
||||||
| NC score | 0.037166 (rank : 96) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.036851 (rank : 97) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
NUPL2_MOUSE
|
||||||
| NC score | 0.036511 (rank : 98) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
NBEA_HUMAN
|
||||||
| NC score | 0.035240 (rank : 99) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
DPOLZ_MOUSE
|
||||||
| NC score | 0.034964 (rank : 100) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
ZCH14_MOUSE
|
||||||
| NC score | 0.034418 (rank : 101) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.032795 (rank : 102) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.032783 (rank : 103) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.032276 (rank : 104) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.031606 (rank : 105) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
MTG8_MOUSE
|
||||||
| NC score | 0.030581 (rank : 106) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.030569 (rank : 107) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
MTG8_HUMAN
|
||||||
| NC score | 0.030547 (rank : 108) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
TR10C_HUMAN
|
||||||
| NC score | 0.030047 (rank : 109) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
IPYR_MOUSE
|
||||||
| NC score | 0.029809 (rank : 110) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D819 | Gene names | Ppa1, Pp, Pyp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase (EC 3.6.1.1) (Pyrophosphate phospho- hydrolase) (PPase). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.029797 (rank : 111) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.029515 (rank : 112) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
TF65_HUMAN
|
||||||
| NC score | 0.028622 (rank : 113) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
PHC2_MOUSE
|
||||||
| NC score | 0.028533 (rank : 114) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
ZN294_HUMAN
|
||||||
| NC score | 0.028522 (rank : 115) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
WNK4_MOUSE
|
||||||
| NC score | 0.027553 (rank : 116) | θ value | 0.00175202 (rank : 5) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
CAF1B_MOUSE
|
||||||
| NC score | 0.027412 (rank : 117) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D0N7 | Gene names | Chaf1b | |||
|
Domain Architecture |

|
|||||
| Description | Chromatin assembly factor 1 subunit B (CAF-1 subunit B) (Chromatin assembly factor I p60 subunit) (CAF-I 60 kDa subunit) (CAF-Ip60). | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.026857 (rank : 118) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.026585 (rank : 119) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CECR6_HUMAN
|
||||||
| NC score | 0.026069 (rank : 120) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXQ6 | Gene names | CECR6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cat eye syndrome critical region protein 6. | |||||
|
APLP1_HUMAN
|
||||||
| NC score | 0.026011 (rank : 121) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
CNTFR_HUMAN
|
||||||
| NC score | 0.025621 (rank : 122) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26992 | Gene names | CNTFR | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary neurotrophic factor receptor alpha precursor (CNTFR alpha). | |||||
|
CNTFR_MOUSE
|
||||||
| NC score | 0.025434 (rank : 123) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88507, Q80T01 | Gene names | Cntfr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary neurotrophic factor receptor alpha precursor (CNTFR alpha). | |||||
|
UBL7_MOUSE
|
||||||
| NC score | 0.025412 (rank : 124) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
UBL7_HUMAN
|
||||||
| NC score | 0.025270 (rank : 125) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96S82, Q96I03 | Gene names | UBL7, BMSCUBP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 7 (Ubiquitin-like protein SB132) (Bone marrow stromal cell ubiquitin-like protein) (BMSC-UbP). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.024762 (rank : 126) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
PLCB1_HUMAN
|
||||||
| NC score | 0.024567 (rank : 127) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
BMP3B_MOUSE
|
||||||
| NC score | 0.023987 (rank : 128) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97737 | Gene names | Gdf10, Bmp3b | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10). | |||||
|
MAGAA_HUMAN
|
||||||
| NC score | 0.023793 (rank : 129) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.023763 (rank : 130) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.023436 (rank : 131) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.022899 (rank : 132) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.022371 (rank : 133) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
FRMD5_HUMAN
|
||||||
| NC score | 0.022001 (rank : 134) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
GP1BA_HUMAN
|
||||||
| NC score | 0.021805 (rank : 135) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.021754 (rank : 136) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
ABI2_HUMAN
|
||||||
| NC score | 0.021481 (rank : 137) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
JADE3_HUMAN
|
||||||
| NC score | 0.021270 (rank : 138) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.021173 (rank : 139) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
CHSTC_HUMAN
|
||||||
| NC score | 0.020999 (rank : 140) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRB3, Q9NXY7 | Gene names | CHST12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2) (Sulfotransferase Hlo). | |||||
|
LCP2_MOUSE
|
||||||
| NC score | 0.020747 (rank : 141) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60787 | Gene names | Lcp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.020564 (rank : 142) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.019638 (rank : 143) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
HOMEZ_HUMAN
|
||||||
| NC score | 0.019416 (rank : 144) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
LYRIC_MOUSE
|
||||||
| NC score | 0.019398 (rank : 145) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
NOP56_MOUSE
|
||||||
| NC score | 0.019342 (rank : 146) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.018240 (rank : 147) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.018131 (rank : 148) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
UBAP1_HUMAN
|
||||||
| NC score | 0.017425 (rank : 149) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ09, Q4V759, Q5T7B3, Q6FI75, Q8NC52, Q8NCG6, Q8NCH9 | Gene names | UBAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 1 (UBAP). | |||||
|
ESR2_MOUSE
|
||||||
| NC score | 0.017260 (rank : 150) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08537, O35635, O70519 | Gene names | Esr2, Estrb, Nr3a2 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor beta (ER-beta). | |||||
|
ESR1_HUMAN
|
||||||
| NC score | 0.017230 (rank : 151) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P03372, Q13511, Q14276, Q9NU51, Q9UDZ7, Q9UIS7 | Gene names | ESR1, ESR, NR3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
ESR1_MOUSE
|
||||||
| NC score | 0.017177 (rank : 152) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19785, Q9JJT5, Q9QY51, Q9QY52 | Gene names | Esr1, Esr, Estr, Estra, Nr3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
PIWL2_HUMAN
|
||||||
| NC score | 0.017175 (rank : 153) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TC59, Q96SW6, Q9NW28 | Gene names | PIWIL2, HILI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
RMI1_HUMAN
|
||||||
| NC score | 0.017122 (rank : 154) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9A7, Q5SQG8, Q5SQG9, Q6P1Q4, Q6PI89, Q7Z6L6 | Gene names | RMI1, C9orf76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RMI1 homolog. | |||||
|
ESR2_HUMAN
|
||||||
| NC score | 0.017114 (rank : 155) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92731, O60608, O60685, O60702, O60703, O75583, O75584, Q86Z31, Q9UEV6, Q9UHD3, Q9UQK9 | Gene names | ESR2, ESTRB, NR3A2 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor beta (ER-beta). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.017022 (rank : 156) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
PROM1_MOUSE
|
||||||
| NC score | 0.016653 (rank : 157) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54990, O35408 | Gene names | Prom1, Prom, Proml1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133 homolog). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.016151 (rank : 158) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
CU013_HUMAN
|
||||||
| NC score | 0.015975 (rank : 159) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
AMZ1_HUMAN
|
||||||
| NC score | 0.015865 (rank : 160) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q400G9, Q8TF51 | Gene names | AMZ1, KIAA1950 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Archaemetzincin-1 (EC 3.-.-.-) (Archeobacterial metalloproteinase-like protein 1). | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.015828 (rank : 161) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
FOXJ3_MOUSE
|
||||||
| NC score | 0.015726 (rank : 162) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.015441 (rank : 163) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
SEM6D_MOUSE
|
||||||
| NC score | 0.014853 (rank : 164) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.014766 (rank : 165) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.014689 (rank : 166) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
SCAP_HUMAN
|
||||||
| NC score | 0.014604 (rank : 167) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.014299 (rank : 168) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
RGS11_HUMAN
|
||||||
| NC score | 0.014183 (rank : 169) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.014051 (rank : 170) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.013466 (rank : 171) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
SKIL_MOUSE
|
||||||
| NC score | 0.013449 (rank : 172) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | 0.012960 (rank : 173) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
USH2A_MOUSE
|
||||||
| NC score | 0.012332 (rank : 174) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
NGN1_HUMAN
|
||||||
| NC score | 0.012140 (rank : 175) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
PPARA_HUMAN
|
||||||
| NC score | 0.010152 (rank : 176) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07869, Q16241, Q92486, Q92689, Q9Y3N1 | Gene names | PPARA, NR1C1, PPAR | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor alpha (PPAR-alpha). | |||||
|
PXR_HUMAN
|
||||||
| NC score | 0.010017 (rank : 177) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75469, Q96AC7, Q9UJ22, Q9UJ23, Q9UJ24, Q9UJ25, Q9UJ26, Q9UJ27 | Gene names | NR1I2, PXR | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor PXR (Pregnane X receptor) (Orphan nuclear receptor PAR1) (Steroid and xenobiotic receptor) (SXR). | |||||
|
K1C18_HUMAN
|
||||||
| NC score | 0.009928 (rank : 178) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05783, Q9BW26 | Gene names | KRT18, CYK18 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 18 (Cytokeratin-18) (CK-18) (Keratin-18) (K18). | |||||
|
EGR3_MOUSE
|
||||||
| NC score | 0.009672 (rank : 179) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43300, Q9R276 | Gene names | Egr3, Egr-3 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 3 (EGR-3). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.009161 (rank : 180) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
CENG2_HUMAN
|
||||||
| NC score | 0.009145 (rank : 181) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
PPP5_HUMAN
|
||||||
| NC score | 0.009134 (rank : 182) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
EGR3_HUMAN
|
||||||
| NC score | 0.008837 (rank : 183) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06889 | Gene names | EGR3, PILOT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 3 (EGR-3) (Zinc finger protein pilot). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.008637 (rank : 184) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.008469 (rank : 185) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
RX_HUMAN
|
||||||
| NC score | 0.008255 (rank : 186) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
K1C18_MOUSE
|
||||||
| NC score | 0.007439 (rank : 187) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05784, Q61766 | Gene names | Krt18, Kerd, Krt1-18 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 18 (Cytokeratin-18) (CK-18) (Keratin-18) (K18) (Cytokeratin endo B) (Keratin D). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.007294 (rank : 188) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
LRC8E_HUMAN
|
||||||
| NC score | 0.005716 (rank : 189) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NSJ5, Q2YDY3, Q7L236, Q8N3B0, Q9H5H8 | Gene names | LRRC8E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8E. | |||||
|
OTX1_MOUSE
|
||||||
| NC score | 0.005500 (rank : 190) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80205 | Gene names | Otx1, Otx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
BAI2_HUMAN
|
||||||
| NC score | 0.005384 (rank : 191) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
DMBT1_HUMAN
|
||||||
| NC score | 0.004958 (rank : 192) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
GUC2C_HUMAN
|
||||||
| NC score | 0.004598 (rank : 193) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.004274 (rank : 194) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
ZN509_HUMAN
|
||||||
| NC score | 0.003496 (rank : 195) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN286_HUMAN
|
||||||
| NC score | 0.002477 (rank : 196) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HBT8, Q96JF3 | Gene names | ZNF286, KIAA1874 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 286. | |||||
|
ZN682_HUMAN
|
||||||
| NC score | 0.001718 (rank : 197) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95780, Q96JV9 | Gene names | ZNF682 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 682. | |||||