Please be patient as the page loads
|
MTG8_HUMAN
|
||||||
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MTG8R_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.970069 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
MTG8R_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.966793 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
MTG8_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
MTG8_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998563 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 5) | NC score | 0.539719 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DEAF1_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 6) | NC score | 0.528834 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
ZMY10_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 7) | NC score | 0.333975 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY10_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 8) | NC score | 0.345720 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99ML0, Q80W73 | Gene names | Zmynd10, Blu | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 9) | NC score | 0.193080 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 10) | NC score | 0.207902 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 11) | NC score | 0.223212 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ANKY2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 12) | NC score | 0.157317 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.220157 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ANKY2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 14) | NC score | 0.161227 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ZMY19_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 15) | NC score | 0.301003 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 16) | NC score | 0.301003 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
THAP7_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.117408 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |
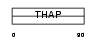
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 18) | NC score | 0.157779 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
EGLN1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.161699 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
KAP1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.050325 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
SMYD3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.182389 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.182039 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
THAP7_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.093349 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |
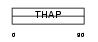
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
PDCD2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.166640 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
PDCD2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.157701 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
ZMY17_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.235855 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
TBX18_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.022135 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |
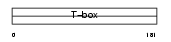
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
RBL2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.032101 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64700 | Gene names | Rbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
ZMY12_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.154386 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.024787 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
SMYD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.144583 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.098543 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
CE016_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.090679 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.021462 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.028237 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NOL7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.065698 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMY1, Q5T297, Q9Y3U7 | Gene names | NOL7, C6orf90, NOP27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 7 (Nucleolar protein of 27 kDa). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.014372 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
KAP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.038191 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.017556 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.019664 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.016142 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.009894 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
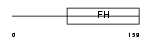
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.016906 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.017404 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.032123 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.016968 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.016445 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KR511_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.034580 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.015405 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.004334 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CAMKV_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.003846 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UHL1, Q8VD20 | Gene names | Camkv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CCD50_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.030941 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q810U5, Q3TRW1, Q8BP82, Q9CZT1, Q9D436 | Gene names | Ccdc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.030547 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.024236 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.016292 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
INP5E_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.014122 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.015117 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
RADI_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.013216 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
SN1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.001659 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57059, Q5R2V5, Q86YJ2 | Gene names | SNF1LK | |||
|
Domain Architecture |
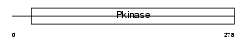
|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK). | |||||
|
ABLM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.004416 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.014896 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RBL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.023037 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08999, Q15073, Q16084, Q92812 | Gene names | RBL2, RB2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.027291 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.022143 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.070042 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SPFH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.025963 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94905, Q6NW21, Q86VS6, Q86W49 | Gene names | SPFH2, C8orf2 | |||
|
Domain Architecture |
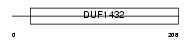
|
|||||
| Description | SPFH domain-containing protein 2 precursor. | |||||
|
SPFH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.025963 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BFZ9, Q8BML8 | Gene names | Spfh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPFH domain-containing protein 2 precursor. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.007905 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
UB2Q2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.018557 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVN8, Q8N4G6, Q96J08 | Gene names | UBE2Q2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 Q2 (EC 6.3.2.19) (Ubiquitin-protein ligase Q2) (Ubiquitin carrier protein Q2). | |||||
|
ZMY15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.059073 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.019537 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.008227 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MBD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.012788 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95243, Q7Z4T3, Q96F09 | Gene names | MBD4, MED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 4 (EC 3.2.2.-) (Methyl-CpG-binding protein MBD4) (Methyl-CpG-binding endonuclease 1) (Mismatch-specific DNA N-glycosylase). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.017122 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SALL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.000540 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.011555 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMYD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.120242 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
TTC14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.015046 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
ABCAD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.005391 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
CING_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.005855 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.013279 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.012291 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.003422 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.006035 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
SMYD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.088015 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
SMYD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.087974 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.011767 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
MTG8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
MTG8_MOUSE
|
||||||
| NC score | 0.998563 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
MTG8R_HUMAN
|
||||||
| NC score | 0.970069 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
MTG8R_MOUSE
|
||||||
| NC score | 0.966793 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.539719 (rank : 5) | θ value | 9.90251e-15 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DEAF1_MOUSE
|
||||||
| NC score | 0.528834 (rank : 6) | θ value | 1.29331e-14 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
ZMY10_MOUSE
|
||||||
| NC score | 0.345720 (rank : 7) | θ value | 1.87187e-05 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99ML0, Q80W73 | Gene names | Zmynd10, Blu | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY10_HUMAN
|
||||||
| NC score | 0.333975 (rank : 8) | θ value | 1.87187e-05 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY19_HUMAN
|
||||||
| NC score | 0.301003 (rank : 9) | θ value | 0.00390308 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| NC score | 0.301003 (rank : 10) | θ value | 0.00390308 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY17_MOUSE
|
||||||
| NC score | 0.235855 (rank : 11) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.223212 (rank : 12) | θ value | 0.00035302 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.220157 (rank : 13) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.207902 (rank : 14) | θ value | 9.29e-05 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.193080 (rank : 15) | θ value | 7.1131e-05 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
SMYD3_HUMAN
|
||||||
| NC score | 0.182389 (rank : 16) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_MOUSE
|
||||||
| NC score | 0.182039 (rank : 17) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
PDCD2_HUMAN
|
||||||
| NC score | 0.166640 (rank : 18) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
EGLN1_HUMAN
|
||||||
| NC score | 0.161699 (rank : 19) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
ANKY2_MOUSE
|
||||||
| NC score | 0.161227 (rank : 20) | θ value | 0.00175202 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.157779 (rank : 21) | θ value | 0.0330416 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
PDCD2_MOUSE
|
||||||
| NC score | 0.157701 (rank : 22) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
ANKY2_HUMAN
|
||||||
| NC score | 0.157317 (rank : 23) | θ value | 0.000786445 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ZMY12_HUMAN
|
||||||
| NC score | 0.154386 (rank : 24) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
SMYD2_HUMAN
|
||||||
| NC score | 0.144583 (rank : 25) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
SMYD2_MOUSE
|
||||||
| NC score | 0.120242 (rank : 26) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
THAP7_HUMAN
|
||||||
| NC score | 0.117408 (rank : 27) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.098543 (rank : 28) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
THAP7_MOUSE
|
||||||
| NC score | 0.093349 (rank : 29) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.090679 (rank : 30) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
SMYD1_HUMAN
|
||||||
| NC score | 0.088015 (rank : 31) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
SMYD1_MOUSE
|
||||||
| NC score | 0.087974 (rank : 32) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.070042 (rank : 33) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
NOL7_HUMAN
|
||||||
| NC score | 0.065698 (rank : 34) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMY1, Q5T297, Q9Y3U7 | Gene names | NOL7, C6orf90, NOP27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 7 (Nucleolar protein of 27 kDa). | |||||
|
ZMY15_HUMAN
|
||||||
| NC score | 0.059073 (rank : 35) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.050325 (rank : 36) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.038191 (rank : 37) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.034580 (rank : 38) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.032123 (rank : 39) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
RBL2_MOUSE
|
||||||
| NC score | 0.032101 (rank : 40) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64700 | Gene names | Rbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
CCD50_MOUSE
|
||||||
| NC score | 0.030941 (rank : 41) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q810U5, Q3TRW1, Q8BP82, Q9CZT1, Q9D436 | Gene names | Ccdc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.030547 (rank : 42) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.028237 (rank : 43) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.027291 (rank : 44) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SPFH2_MOUSE
|
||||||
| NC score | 0.025963 (rank : 45) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BFZ9, Q8BML8 | Gene names | Spfh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPFH domain-containing protein 2 precursor. | |||||
|
SPFH2_HUMAN
|
||||||
| NC score | 0.025963 (rank : 46) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94905, Q6NW21, Q86VS6, Q86W49 | Gene names | SPFH2, C8orf2 | |||
|
Domain Architecture |

|
|||||
| Description | SPFH domain-containing protein 2 precursor. | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.024787 (rank : 47) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.024236 (rank : 48) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
RBL2_HUMAN
|
||||||
| NC score | 0.023037 (rank : 49) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08999, Q15073, Q16084, Q92812 | Gene names | RBL2, RB2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.022143 (rank : 50) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TBX18_HUMAN
|
||||||
| NC score | 0.022135 (rank : 51) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.021462 (rank : 52) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.019664 (rank : 53) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.019537 (rank : 54) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
UB2Q2_HUMAN
|
||||||
| NC score | 0.018557 (rank : 55) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVN8, Q8N4G6, Q96J08 | Gene names | UBE2Q2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 Q2 (EC 6.3.2.19) (Ubiquitin-protein ligase Q2) (Ubiquitin carrier protein Q2). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.017556 (rank : 56) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.017404 (rank : 57) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.017122 (rank : 58) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.016968 (rank : 59) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.016906 (rank : 60) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.016445 (rank : 61) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.016292 (rank : 62) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.016142 (rank : 63) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.015405 (rank : 64) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.015117 (rank : 65) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.015046 (rank : 66) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.014896 (rank : 67) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.014372 (rank : 68) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
INP5E_HUMAN
|
||||||
| NC score | 0.014122 (rank : 69) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.013279 (rank : 70) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.013216 (rank : 71) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
MBD4_HUMAN
|
||||||
| NC score | 0.012788 (rank : 72) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95243, Q7Z4T3, Q96F09 | Gene names | MBD4, MED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 4 (EC 3.2.2.-) (Methyl-CpG-binding protein MBD4) (Methyl-CpG-binding endonuclease 1) (Mismatch-specific DNA N-glycosylase). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.012291 (rank : 73) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.011767 (rank : 74) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.011555 (rank : 75) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.009894 (rank : 76) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.008227 (rank : 77) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.007905 (rank : 78) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.006035 (rank : 79) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.005855 (rank : 80) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
ABCAD_HUMAN
|
||||||
| NC score | 0.005391 (rank : 81) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
ABLM1_MOUSE
|
||||||
| NC score | 0.004416 (rank : 82) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.004334 (rank : 83) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CAMKV_MOUSE
|
||||||
| NC score | 0.003846 (rank : 84) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UHL1, Q8VD20 | Gene names | Camkv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.003422 (rank : 85) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
SN1L1_HUMAN
|
||||||
| NC score | 0.001659 (rank : 86) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57059, Q5R2V5, Q86YJ2 | Gene names | SNF1LK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK). | |||||
|
SALL1_MOUSE
|
||||||
| NC score | 0.000540 (rank : 87) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||