Please be patient as the page loads
|
PDCD2_MOUSE
|
||||||
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDCD2_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
PDCD2_HUMAN
|
||||||
| θ value | 1.6163e-166 (rank : 2) | NC score | 0.994321 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
EGLN1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 3) | NC score | 0.207466 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 4) | NC score | 0.193714 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
ZMY12_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 5) | NC score | 0.227642 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
SMYD2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 6) | NC score | 0.166839 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.122095 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.159910 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
DEAF1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.160105 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
MTG8_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.157701 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
MTG8_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.156296 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
TESK2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.010624 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |
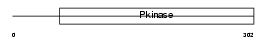
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
MTG8R_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.152383 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.156460 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
MTG8R_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.146864 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
SMYD1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.113314 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
ZMY10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.147825 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY10_MOUSE
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.147405 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99ML0, Q80W73 | Gene names | Zmynd10, Blu | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY17_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.168780 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
SMYD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.105540 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.117282 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
ZMY19_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.113015 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.113015 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.003705 (rank : 34) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CHIC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.018494 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VXU3 | Gene names | CHIC1, BRX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
NBEAL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.011746 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
TEX9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.009933 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.004231 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
SMYD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.103449 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
EGLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.070405 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KS0, Q8WWY4, Q9BV14 | Gene names | EGLN2, EIT6 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 2 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 1) (HIF-prolyl hydroxylase 1) (HIF-PH1) (HPH-3) (Prolyl hydroxylase domain-containing protein 1) (PHD1) (Estrogen-induced tag 6). | |||||
|
EGLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.069702 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YE2, Q8C6I4, Q8CIL9, Q8VHJ1, Q99MI0 | Gene names | Egln2 | |||
|
Domain Architecture |
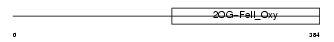
|
|||||
| Description | Egl nine homolog 2 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 1) (HIF-prolyl hydroxylase 1) (HIF-PH1) (Falkor). | |||||
|
EGLN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.072637 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H6Z9 | Gene names | EGLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (HPH-1) (Prolyl hydroxylase domain-containing protein 3) (PHD3). | |||||
|
EGLN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.072402 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91UZ4, Q3TCG8, Q8C8M6, Q8CCA8, Q8R5C7 | Gene names | Egln3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (SM-20). | |||||
|
SMYD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.096473 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
PDCD2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
PDCD2_HUMAN
|
||||||
| NC score | 0.994321 (rank : 2) | θ value | 1.6163e-166 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
ZMY12_HUMAN
|
||||||
| NC score | 0.227642 (rank : 3) | θ value | 0.0252991 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
EGLN1_HUMAN
|
||||||
| NC score | 0.207466 (rank : 4) | θ value | 0.00020696 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.193714 (rank : 5) | θ value | 0.00298849 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
ZMY17_MOUSE
|
||||||
| NC score | 0.168780 (rank : 6) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
SMYD2_MOUSE
|
||||||
| NC score | 0.166839 (rank : 7) | θ value | 0.125558 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
DEAF1_MOUSE
|
||||||
| NC score | 0.160105 (rank : 8) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
SMYD2_HUMAN
|
||||||
| NC score | 0.159910 (rank : 9) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
MTG8_HUMAN
|
||||||
| NC score | 0.157701 (rank : 10) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.156460 (rank : 11) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
MTG8_MOUSE
|
||||||
| NC score | 0.156296 (rank : 12) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
MTG8R_MOUSE
|
||||||
| NC score | 0.152383 (rank : 13) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
ZMY10_HUMAN
|
||||||
| NC score | 0.147825 (rank : 14) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY10_MOUSE
|
||||||
| NC score | 0.147405 (rank : 15) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99ML0, Q80W73 | Gene names | Zmynd10, Blu | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
MTG8R_HUMAN
|
||||||
| NC score | 0.146864 (rank : 16) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.122095 (rank : 17) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.117282 (rank : 18) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD1_MOUSE
|
||||||
| NC score | 0.113314 (rank : 19) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
ZMY19_HUMAN
|
||||||
| NC score | 0.113015 (rank : 20) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| NC score | 0.113015 (rank : 21) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
SMYD1_HUMAN
|
||||||
| NC score | 0.105540 (rank : 22) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
SMYD3_HUMAN
|
||||||
| NC score | 0.103449 (rank : 23) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_MOUSE
|
||||||
| NC score | 0.096473 (rank : 24) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
EGLN3_HUMAN
|
||||||
| NC score | 0.072637 (rank : 25) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H6Z9 | Gene names | EGLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (HPH-1) (Prolyl hydroxylase domain-containing protein 3) (PHD3). | |||||
|
EGLN3_MOUSE
|
||||||
| NC score | 0.072402 (rank : 26) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91UZ4, Q3TCG8, Q8C8M6, Q8CCA8, Q8R5C7 | Gene names | Egln3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (SM-20). | |||||
|
EGLN2_HUMAN
|
||||||
| NC score | 0.070405 (rank : 27) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KS0, Q8WWY4, Q9BV14 | Gene names | EGLN2, EIT6 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 2 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 1) (HIF-prolyl hydroxylase 1) (HIF-PH1) (HPH-3) (Prolyl hydroxylase domain-containing protein 1) (PHD1) (Estrogen-induced tag 6). | |||||
|
EGLN2_MOUSE
|
||||||
| NC score | 0.069702 (rank : 28) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YE2, Q8C6I4, Q8CIL9, Q8VHJ1, Q99MI0 | Gene names | Egln2 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 2 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 1) (HIF-prolyl hydroxylase 1) (HIF-PH1) (Falkor). | |||||
|
CHIC1_HUMAN
|
||||||
| NC score | 0.018494 (rank : 29) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VXU3 | Gene names | CHIC1, BRX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
NBEAL_HUMAN
|
||||||
| NC score | 0.011746 (rank : 30) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
TESK2_MOUSE
|
||||||
| NC score | 0.010624 (rank : 31) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TEX9_MOUSE
|
||||||
| NC score | 0.009933 (rank : 32) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.004231 (rank : 33) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.003705 (rank : 34) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||