Please be patient as the page loads
|
MTG8R_HUMAN
|
||||||
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MTG8R_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
MTG8R_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991289 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
MTG8_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.970069 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
MTG8_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.971069 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 5) | NC score | 0.501198 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DEAF1_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 6) | NC score | 0.488723 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 7) | NC score | 0.204457 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ANKY2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 8) | NC score | 0.158567 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ANKY2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 9) | NC score | 0.162316 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ZMY10_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 10) | NC score | 0.312053 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99ML0, Q80W73 | Gene names | Zmynd10, Blu | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY19_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 11) | NC score | 0.310119 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 12) | NC score | 0.310119 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY10_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 13) | NC score | 0.299623 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
SMYD3_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.196684 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.196658 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
EGLN1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.161105 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.170700 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.156288 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.034696 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.111968 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
PRP6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.058199 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94906, O95109, Q5VXS5, Q9H3Z1, Q9H4T9, Q9H4U8, Q9NTE6 | Gene names | PRPF6, C20orf14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
PRP6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.058096 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YR7, Q8CIK9, Q8R3M8, Q99JN1, Q9CSZ0 | Gene names | Prpf6 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
SMYD2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.158441 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.169442 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.172905 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.044128 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ZMY12_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.159795 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
CE016_HUMAN
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.097653 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
ERRFI_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.064411 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.025634 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.042002 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.030899 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.063631 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
ZMY17_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.225891 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
HES7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.057165 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.030929 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.030505 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.029933 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PDCD2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.154450 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.032927 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.053401 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.051150 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.041177 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
NC6IP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.035149 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
PDCD2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.146864 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.056389 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.029390 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.053209 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.037495 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
KLC4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.035803 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.016300 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.032101 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.040207 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.035920 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.033865 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.042232 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
FRS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.028315 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
HXD3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.014219 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
KLC4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.033264 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
MYST1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.028043 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D1P2, Q3UIY0, Q8BJ69, Q8BJ76, Q8CI73 | Gene names | Myst1, Mof | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable histone acetyltransferase MYST1 (EC 2.3.1.48) (MYST protein 1) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 1). | |||||
|
FOXJ3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.011632 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.029998 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.026745 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.025020 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
SMYD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.133463 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
SO4A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.015843 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96BD0, Q9H4T7, Q9H4T8, Q9H8P2, Q9NWX8, Q9UI35, Q9UIG7 | Gene names | SLCO4A1, OATP1, OATPE, SLC21A12 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 4A1 (Solute carrier family 21 member 12) (Sodium-independent organic anion transporter E) (Organic anion-transporting polypeptide E) (OATP-E) (Colon organic anion transporter) (Organic anion transporter polypeptide-related protein 1) (OATP-RP1) (OATPRP1) (POAT). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.028246 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.022112 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.030438 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.009578 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.034854 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.010924 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.030869 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
ERRFI_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.043280 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
ESX1L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.011824 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
FA50A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.022437 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WV03, Q9D866 | Gene names | Fam50a, D0HXS9928E, Xap5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein FAM50A (XAP-5 protein). | |||||
|
GAH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.030118 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63118 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_3q26 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.019123 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
INP4B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.019578 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15327, Q6IN59, Q6PJB4 | Gene names | INPP4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type II). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.028892 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.001906 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
RASF8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.022740 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NHQ8, O95647, Q5SCI2, Q76KB6 | Gene names | RASSF8, C12orf2 | |||
|
Domain Architecture |
|
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.019915 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.081763 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.020123 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
UB7I1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.047960 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.031149 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.010033 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.031637 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.027759 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.019689 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
HXD3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.010433 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
LNP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.018819 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.006146 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.028983 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.029159 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.016034 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.030522 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SFR17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.015922 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.026751 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.014872 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
ZMY15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.059382 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.009321 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.029588 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
DREB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.016726 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
FA50A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.018068 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14320, Q5HY37 | Gene names | FAM50A, DXS9928E, HXC26, XAP5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein FAM50A (Protein XAP-5) (Protein HXC-26). | |||||
|
K1C16_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.009485 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2K1 | Gene names | Krt16, Krt1-16 | |||
|
Domain Architecture |
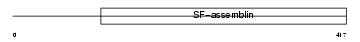
|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
LPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.008436 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
NMT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.010004 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70310 | Gene names | Nmt1 | |||
|
Domain Architecture |
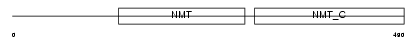
|
|||||
| Description | Glycylpeptide N-tetradecanoyltransferase 1 (EC 2.3.1.97) (Peptide N- myristoyltransferase 1) (Myristoyl-CoA:protein N-myristoyltransferase 1) (NMT 1) (Type I N-myristoyltransferase). | |||||
|
SMYD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.102018 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
SMYD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.101600 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.001477 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.014064 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TRI41_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.003493 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WV44, Q5BKT0, Q7L484, Q96Q10, Q9BSL8 | Gene names | TRIM41 | |||
|
Domain Architecture |
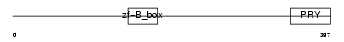
|
|||||
| Description | Tripartite motif-containing protein 41. | |||||
|
TXLNB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.016205 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.024070 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CUED1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.012008 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWM3, Q9NWD0 | Gene names | CUEDC1 | |||
|
Domain Architecture |
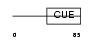
|
|||||
| Description | CUE domain-containing protein 1. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.016887 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KR511_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.031633 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.018805 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
NOG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.022243 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZE4, O95446, Q9NVJ8 | Gene names | GTPBP4, CRFG, NOG1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
PRRX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.004267 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06348, Q61651 | Gene names | Prrx2, Prx2, S8 | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2 (PRX-2) (Homeobox protein S8). | |||||
|
RB15B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.005983 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RBL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.016632 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64700 | Gene names | Rbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.014258 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
THAP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.059748 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
MTG8R_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
MTG8R_MOUSE
|
||||||
| NC score | 0.991289 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
MTG8_MOUSE
|
||||||
| NC score | 0.971069 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
MTG8_HUMAN
|
||||||
| NC score | 0.970069 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.501198 (rank : 5) | θ value | 2.28291e-11 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DEAF1_MOUSE
|
||||||
| NC score | 0.488723 (rank : 6) | θ value | 2.52405e-10 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
ZMY10_MOUSE
|
||||||
| NC score | 0.312053 (rank : 7) | θ value | 0.000602161 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99ML0, Q80W73 | Gene names | Zmynd10, Blu | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY19_HUMAN
|
||||||
| NC score | 0.310119 (rank : 8) | θ value | 0.000786445 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| NC score | 0.310119 (rank : 9) | θ value | 0.000786445 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY10_HUMAN
|
||||||
| NC score | 0.299623 (rank : 10) | θ value | 0.00390308 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY17_MOUSE
|
||||||
| NC score | 0.225891 (rank : 11) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.204457 (rank : 12) | θ value | 1.43324e-05 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
SMYD3_MOUSE
|
||||||
| NC score | 0.196684 (rank : 13) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_HUMAN
|
||||||
| NC score | 0.196658 (rank : 14) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.172905 (rank : 15) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.170700 (rank : 16) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.169442 (rank : 17) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ANKY2_MOUSE
|
||||||
| NC score | 0.162316 (rank : 18) | θ value | 0.000461057 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
EGLN1_HUMAN
|
||||||
| NC score | 0.161105 (rank : 19) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
ZMY12_HUMAN
|
||||||
| NC score | 0.159795 (rank : 20) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
ANKY2_HUMAN
|
||||||
| NC score | 0.158567 (rank : 21) | θ value | 0.000461057 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
SMYD2_HUMAN
|
||||||
| NC score | 0.158441 (rank : 22) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.156288 (rank : 23) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
PDCD2_HUMAN
|
||||||
| NC score | 0.154450 (rank : 24) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
PDCD2_MOUSE
|
||||||
| NC score | 0.146864 (rank : 25) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
SMYD2_MOUSE
|
||||||
| NC score | 0.133463 (rank : 26) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.111968 (rank : 27) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD1_HUMAN
|
||||||
| NC score | 0.102018 (rank : 28) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
SMYD1_MOUSE
|
||||||
| NC score | 0.101600 (rank : 29) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.097653 (rank : 30) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.081763 (rank : 31) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
ERRFI_HUMAN
|
||||||
| NC score | 0.064411 (rank : 32) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.063631 (rank : 33) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
THAP7_HUMAN
|
||||||
| NC score | 0.059748 (rank : 34) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
ZMY15_HUMAN
|
||||||
| NC score | 0.059382 (rank : 35) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
PRP6_HUMAN
|
||||||
| NC score | 0.058199 (rank : 36) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94906, O95109, Q5VXS5, Q9H3Z1, Q9H4T9, Q9H4U8, Q9NTE6 | Gene names | PRPF6, C20orf14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
PRP6_MOUSE
|
||||||
| NC score | 0.058096 (rank : 37) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YR7, Q8CIK9, Q8R3M8, Q99JN1, Q9CSZ0 | Gene names | Prpf6 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.057165 (rank : 38) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.056389 (rank : 39) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.053401 (rank : 40) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.053209 (rank : 41) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.051150 (rank : 42) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
UB7I1_HUMAN
|
||||||
| NC score | 0.047960 (rank : 43) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.044128 (rank : 44) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ERRFI_MOUSE
|
||||||
| NC score | 0.043280 (rank : 45) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.042232 (rank : 46) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.042002 (rank : 47) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.041177 (rank : 48) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.040207 (rank : 49) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.037495 (rank : 50) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.035920 (rank : 51) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
KLC4_MOUSE
|
||||||
| NC score | 0.035803 (rank : 52) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
NC6IP_MOUSE
|
||||||
| NC score | 0.035149 (rank : 53) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.034854 (rank : 54) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.034696 (rank : 55) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.033865 (rank : 56) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.033264 (rank : 57) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.032927 (rank : 58) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.032101 (rank : 59) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.031637 (rank : 60) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.031633 (rank : 61) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.031149 (rank : 62) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.030929 (rank : 63) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.030899 (rank : 64) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.030869 (rank : 65) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.030522 (rank : 66) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.030505 (rank : 67) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.030438 (rank : 68) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
GAH2_HUMAN
|
||||||
| NC score | 0.030118 (rank : 69) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63118 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_3q26 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.029998 (rank : 70) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.029933 (rank : 71) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.029588 (rank : 72) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.029390 (rank : 73) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.029159 (rank : 74) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.028983 (rank : 75) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.028892 (rank : 76) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
FRS3_MOUSE
|
||||||
| NC score | 0.028315 (rank : 77) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.028246 (rank : 78) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MYST1_MOUSE
|
||||||
| NC score | 0.028043 (rank : 79) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D1P2, Q3UIY0, Q8BJ69, Q8BJ76, Q8CI73 | Gene names | Myst1, Mof | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable histone acetyltransferase MYST1 (EC 2.3.1.48) (MYST protein 1) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 1). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.027759 (rank : 80) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.026751 (rank : 81) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.026745 (rank : 82) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.025634 (rank : 83) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.025020 (rank : 84) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.024070 (rank : 85) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
RASF8_HUMAN
|
||||||
| NC score | 0.022740 (rank : 86) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NHQ8, O95647, Q5SCI2, Q76KB6 | Gene names | RASSF8, C12orf2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1). | |||||
|
FA50A_MOUSE
|
||||||
| NC score | 0.022437 (rank : 87) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WV03, Q9D866 | Gene names | Fam50a, D0HXS9928E, Xap5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM50A (XAP-5 protein). | |||||
|
NOG1_HUMAN
|
||||||
| NC score | 0.022243 (rank : 88) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZE4, O95446, Q9NVJ8 | Gene names | GTPBP4, CRFG, NOG1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.022112 (rank : 89) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.020123 (rank : 90) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.019915 (rank : 91) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.019689 (rank : 92) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
INP4B_HUMAN
|
||||||
| NC score | 0.019578 (rank : 93) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15327, Q6IN59, Q6PJB4 | Gene names | INPP4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type II). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.019123 (rank : 94) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
LNP_MOUSE
|
||||||
| NC score | 0.018819 (rank : 95) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.018805 (rank : 96) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
FA50A_HUMAN
|
||||||
| NC score | 0.018068 (rank : 97) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14320, Q5HY37 | Gene names | FAM50A, DXS9928E, HXC26, XAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM50A (Protein XAP-5) (Protein HXC-26). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.016887 (rank : 98) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
DREB_HUMAN
|
||||||
| NC score | 0.016726 (rank : 99) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
RBL2_MOUSE
|
||||||
| NC score | 0.016632 (rank : 100) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64700 | Gene names | Rbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.016300 (rank : 101) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
TXLNB_MOUSE
|
||||||
| NC score | 0.016205 (rank : 102) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.016034 (rank : 103) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.015922 (rank : 104) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
SO4A1_HUMAN
|
||||||
| NC score | 0.015843 (rank : 105) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96BD0, Q9H4T7, Q9H4T8, Q9H8P2, Q9NWX8, Q9UI35, Q9UIG7 | Gene names | SLCO4A1, OATP1, OATPE, SLC21A12 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 4A1 (Solute carrier family 21 member 12) (Sodium-independent organic anion transporter E) (Organic anion-transporting polypeptide E) (OATP-E) (Colon organic anion transporter) (Organic anion transporter polypeptide-related protein 1) (OATP-RP1) (OATPRP1) (POAT). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.014872 (rank : 106) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.014258 (rank : 107) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
HXD3_HUMAN
|
||||||
| NC score | 0.014219 (rank : 108) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.014064 (rank : 109) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
CUED1_HUMAN
|
||||||
| NC score | 0.012008 (rank : 110) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWM3, Q9NWD0 | Gene names | CUEDC1 | |||
|
Domain Architecture |

|
|||||
| Description | CUE domain-containing protein 1. | |||||
|
ESX1L_HUMAN
|
||||||
| NC score | 0.011824 (rank : 111) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
FOXJ3_HUMAN
|
||||||
| NC score | 0.011632 (rank : 112) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.010924 (rank : 113) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
HXD3_MOUSE
|
||||||
| NC score | 0.010433 (rank : 114) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.010033 (rank : 115) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
NMT1_MOUSE
|
||||||
| NC score | 0.010004 (rank : 116) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70310 | Gene names | Nmt1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycylpeptide N-tetradecanoyltransferase 1 (EC 2.3.1.97) (Peptide N- myristoyltransferase 1) (Myristoyl-CoA:protein N-myristoyltransferase 1) (NMT 1) (Type I N-myristoyltransferase). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.009578 (rank : 117) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
K1C16_MOUSE
|
||||||
| NC score | 0.009485 (rank : 118) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2K1 | Gene names | Krt16, Krt1-16 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.009321 (rank : 119) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
LPP_MOUSE
|
||||||
| NC score | 0.008436 (rank : 120) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.006146 (rank : 121) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
RB15B_HUMAN
|
||||||
| NC score | 0.005983 (rank : 122) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
PRRX2_MOUSE
|
||||||
| NC score | 0.004267 (rank : 123) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06348, Q61651 | Gene names | Prrx2, Prx2, S8 | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2 (PRX-2) (Homeobox protein S8). | |||||
|
TRI41_HUMAN
|
||||||
| NC score | 0.003493 (rank : 124) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WV44, Q5BKT0, Q7L484, Q96Q10, Q9BSL8 | Gene names | TRIM41 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 41. | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.001906 (rank : 125) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.001477 (rank : 126) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||