Please be patient as the page loads
|
ERRFI_HUMAN
|
||||||
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ERRFI_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
ERRFI_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.976022 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
ACK1_MOUSE
|
||||||
| θ value | 1.2049e-28 (rank : 3) | NC score | 0.138348 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |
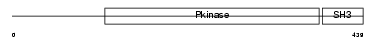
|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
ACK1_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 4) | NC score | 0.137642 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.120255 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
ATX1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.093937 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54253, Q9UJG2, Q9Y4J1 | Gene names | ATXN1, ATX1, SCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.079215 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
SALL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.014502 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
SNPC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.069510 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.108238 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.088383 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.064643 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
MTG8R_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.064411 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.039529 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MTG8R_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.059416 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
ZN645_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.074389 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.030060 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.014120 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.065895 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.050814 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.085612 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.054984 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.047633 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
SALL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.013450 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||
|
TBC17_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.027050 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HA65 | Gene names | TBC1D17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 17. | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.033309 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DTX1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.029747 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |
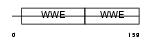
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
FOXP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.017717 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
NECD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.021041 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |
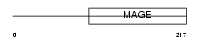
|
|||||
| Description | Necdin. | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.068377 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.041461 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.052503 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.029186 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.032497 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.056160 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.050251 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.038660 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.011841 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.039501 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
SETB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.024799 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.018353 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.018767 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
CF055_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.054104 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP79, Q9H0R2, Q9H3K9, Q9P0Q0 | Gene names | C6orf55 | |||
|
Domain Architecture |
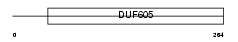
|
|||||
| Description | Protein C6orf55 (Dopamine-responsive protein DRG-1). | |||||
|
ESAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.019806 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925F2 | Gene names | Esam, Esam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.038788 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
AL2SB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.036993 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96Q35 | Gene names | ALS2CR12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 12 protein. | |||||
|
CD248_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.026335 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.033903 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.032707 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.033634 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.065738 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.007703 (rank : 77) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
RFIP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.017145 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
RTN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.045589 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.025187 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CALX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.022483 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
DMXL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.010830 (rank : 75) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.009443 (rank : 76) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.041913 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.047116 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.022702 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.011160 (rank : 74) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.044857 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN750_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.062494 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.011593 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
CMTA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.014519 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
DAB2P_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.014033 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
EDAR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.013489 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
EVL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.020193 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
IF16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.013794 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.015961 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ZP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.011548 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60852 | Gene names | ZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida sperm-binding protein 1 precursor (Zona pellucida glycoprotein 1) (Zp-1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.055605 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.050353 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TNK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.051330 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13470, O95364, Q8IYI4 | Gene names | TNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (CD38 negative kinase 1). | |||||
|
TNK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.050413 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99ML2, Q8CCC4, Q8K0X9, Q91V11 | Gene names | Tnk1, Kos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (Kinase of embryonic stem cells). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.052851 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
ERRFI_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
ERRFI_MOUSE
|
||||||
| NC score | 0.976022 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
ACK1_MOUSE
|
||||||
| NC score | 0.138348 (rank : 3) | θ value | 1.2049e-28 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
ACK1_HUMAN
|
||||||
| NC score | 0.137642 (rank : 4) | θ value | 5.97985e-28 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.120255 (rank : 5) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.108238 (rank : 6) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
ATX1_HUMAN
|
||||||
| NC score | 0.093937 (rank : 7) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54253, Q9UJG2, Q9Y4J1 | Gene names | ATXN1, ATX1, SCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.088383 (rank : 8) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.085612 (rank : 9) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.079215 (rank : 10) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZN645_HUMAN
|
||||||
| NC score | 0.074389 (rank : 11) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
SNPC2_HUMAN
|
||||||
| NC score | 0.069510 (rank : 12) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.068377 (rank : 13) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.065895 (rank : 14) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.065738 (rank : 15) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.064643 (rank : 16) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
MTG8R_HUMAN
|
||||||
| NC score | 0.064411 (rank : 17) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
ZN750_HUMAN
|
||||||
| NC score | 0.062494 (rank : 18) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
MTG8R_MOUSE
|
||||||
| NC score | 0.059416 (rank : 19) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.056160 (rank : 20) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.055605 (rank : 21) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.054984 (rank : 22) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CF055_HUMAN
|
||||||
| NC score | 0.054104 (rank : 23) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP79, Q9H0R2, Q9H3K9, Q9P0Q0 | Gene names | C6orf55 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C6orf55 (Dopamine-responsive protein DRG-1). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.052851 (rank : 24) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.052503 (rank : 25) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TNK1_HUMAN
|
||||||
| NC score | 0.051330 (rank : 26) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13470, O95364, Q8IYI4 | Gene names | TNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (CD38 negative kinase 1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.050814 (rank : 27) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TNK1_MOUSE
|
||||||
| NC score | 0.050413 (rank : 28) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99ML2, Q8CCC4, Q8K0X9, Q91V11 | Gene names | Tnk1, Kos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (Kinase of embryonic stem cells). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.050353 (rank : 29) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.050251 (rank : 30) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.047633 (rank : 31) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.047116 (rank : 32) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RTN4_HUMAN
|
||||||
| NC score | 0.045589 (rank : 33) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.044857 (rank : 34) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.041913 (rank : 35) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.041461 (rank : 36) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.039529 (rank : 37) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.039501 (rank : 38) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.038788 (rank : 39) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.038660 (rank : 40) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
AL2SB_HUMAN
|
||||||
| NC score | 0.036993 (rank : 41) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96Q35 | Gene names | ALS2CR12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 12 protein. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.033903 (rank : 42) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.033634 (rank : 43) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.033309 (rank : 44) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.032707 (rank : 45) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.032497 (rank : 46) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.030060 (rank : 47) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
DTX1_MOUSE
|
||||||
| NC score | 0.029747 (rank : 48) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.029186 (rank : 49) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
TBC17_HUMAN
|
||||||
| NC score | 0.027050 (rank : 50) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HA65 | Gene names | TBC1D17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 17. | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.026335 (rank : 51) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.025187 (rank : 52) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SETB1_MOUSE
|
||||||
| NC score | 0.024799 (rank : 53) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.022702 (rank : 54) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.022483 (rank : 55) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
NECD_HUMAN
|
||||||
| NC score | 0.021041 (rank : 56) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.020193 (rank : 57) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
ESAM_MOUSE
|
||||||
| NC score | 0.019806 (rank : 58) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925F2 | Gene names | Esam, Esam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.018767 (rank : 59) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.018353 (rank : 60) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
FOXP4_HUMAN
|
||||||
| NC score | 0.017717 (rank : 61) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
RFIP4_HUMAN
|
||||||
| NC score | 0.017145 (rank : 62) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.015961 (rank : 63) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
CMTA1_HUMAN
|
||||||
| NC score | 0.014519 (rank : 64) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
SALL1_HUMAN
|
||||||
| NC score | 0.014502 (rank : 65) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.014120 (rank : 66) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
DAB2P_HUMAN
|
||||||
| NC score | 0.014033 (rank : 67) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.013794 (rank : 68) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
EDAR_MOUSE
|
||||||
| NC score | 0.013489 (rank : 69) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
SALL1_MOUSE
|
||||||
| NC score | 0.013450 (rank : 70) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.011841 (rank : 71) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.011593 (rank : 72) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZP1_HUMAN
|
||||||
| NC score | 0.011548 (rank : 73) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60852 | Gene names | ZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida sperm-binding protein 1 precursor (Zona pellucida glycoprotein 1) (Zp-1). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.011160 (rank : 74) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
DMXL1_HUMAN
|
||||||
| NC score | 0.010830 (rank : 75) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.009443 (rank : 76) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.007703 (rank : 77) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||