Please be patient as the page loads
|
CALX_HUMAN
|
||||||
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CALX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CALX_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991095 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CLGN_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.972220 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor. | |||||
|
CLGN_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.966992 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
CALR_HUMAN
|
||||||
| θ value | 9.76566e-63 (rank : 5) | NC score | 0.890290 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
CALR_MOUSE
|
||||||
| θ value | 2.17557e-62 (rank : 6) | NC score | 0.892789 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P14211 | Gene names | Calr | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60). | |||||
|
CALR3_HUMAN
|
||||||
| θ value | 1.9192e-34 (rank : 7) | NC score | 0.822332 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L12, Q96LN3 | Gene names | CALR3, CRT2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR3_MOUSE
|
||||||
| θ value | 3.3877e-31 (rank : 8) | NC score | 0.811973 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.106446 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 10) | NC score | 0.090060 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
H6ST3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.079186 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.069897 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.017081 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.069186 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.037488 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.095103 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.036551 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.031274 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.055291 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.038700 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.055321 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.058882 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.039437 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.066019 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
DMXL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.020321 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.055024 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
ACK1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.007120 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.060516 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.066859 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.011069 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.105207 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.035272 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
VSX1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.016143 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
CP39A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.031758 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKJ9 | Gene names | Cyp39a1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (mCYP39A1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.055488 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.025335 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
ACK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.006186 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |
|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.060248 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K1143_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.035499 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K039, Q9CSR4, Q9D0W8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1143 homolog. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.071588 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.030545 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.053587 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.034875 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
PEG3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.005873 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9GZU2, P78418, Q5H9P9, Q7Z7H7, Q8TF75, Q9GZY2 | Gene names | PEG3, KIAA0287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.008076 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.021635 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.030545 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
IRX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.022090 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.052365 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.004840 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.018352 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.025514 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.025435 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
K0460_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.014692 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.010878 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.010679 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.059838 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TSH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.011938 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.003793 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.021822 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.011033 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.008275 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MCES_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.029707 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.044150 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.016290 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.011289 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.021680 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
CCDC9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.058061 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.042498 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CF010_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.054627 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.034449 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CP39A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.017426 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |
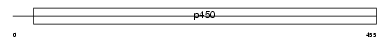
|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
ERRFI_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.022483 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.014272 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
LRMP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.013024 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12912, Q8N301 | Gene names | LRMP, JAW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.019296 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NOL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.024495 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |
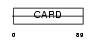
|
|||||
| Description | Nucleolar protein 3. | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.011784 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.009857 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PER2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.006603 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PSMD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.010833 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
TNNT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.018728 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P13805, O95472, Q16061 | Gene names | TNNT1, TNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, slow skeletal muscle (TnTs) (Slow skeletal muscle troponin T) (sTnT). | |||||
|
ASXL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.014305 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.005486 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.032005 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.013458 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.031655 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.008027 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
ENPL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.008516 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08113, P11427 | Gene names | Hsp90b1, Tra-1, Tra1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (ERP99) (Polymorphic tumor rejection antigen 1) (Tumor rejection antigen gp96). | |||||
|
RBBP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.018254 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |
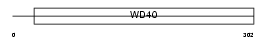
|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.034484 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
SH23A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.006369 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRG2, Q9Y2X4 | Gene names | SH2D3A, NSP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3A (Novel SH2-containing protein 1). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.003606 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.002811 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.003425 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.019893 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.053486 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.072670 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.058428 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.058121 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CALX_MOUSE
|
||||||
| NC score | 0.991095 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CLGN_HUMAN
|
||||||
| NC score | 0.972220 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor. | |||||
|
CLGN_MOUSE
|
||||||
| NC score | 0.966992 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
CALR_MOUSE
|
||||||
| NC score | 0.892789 (rank : 5) | θ value | 2.17557e-62 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P14211 | Gene names | Calr | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60). | |||||
|
CALR_HUMAN
|
||||||
| NC score | 0.890290 (rank : 6) | θ value | 9.76566e-63 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
CALR3_HUMAN
|
||||||
| NC score | 0.822332 (rank : 7) | θ value | 1.9192e-34 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L12, Q96LN3 | Gene names | CALR3, CRT2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR3_MOUSE
|
||||||
| NC score | 0.811973 (rank : 8) | θ value | 3.3877e-31 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.106446 (rank : 9) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.105207 (rank : 10) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.095103 (rank : 11) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.090060 (rank : 12) | θ value | 0.0148317 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
H6ST3_MOUSE
|
||||||
| NC score | 0.079186 (rank : 13) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.072670 (rank : 14) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.071588 (rank : 15) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.069897 (rank : 16) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.069186 (rank : 17) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.066859 (rank : 18) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.066019 (rank : 19) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.060516 (rank : 20) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.060248 (rank : 21) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.059838 (rank : 22) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.058882 (rank : 23) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.058428 (rank : 24) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.058121 (rank : 25) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
CCDC9_HUMAN
|
||||||
| NC score | 0.058061 (rank : 26) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.055488 (rank : 27) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.055321 (rank : 28) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.055291 (rank : 29) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.055024 (rank : 30) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.054627 (rank : 31) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.053587 (rank : 32) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.053486 (rank : 33) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.052365 (rank : 34) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.044150 (rank : 35) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.042498 (rank : 36) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.039437 (rank : 37) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.038700 (rank : 38) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.037488 (rank : 39) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.036551 (rank : 40) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
K1143_MOUSE
|
||||||
| NC score | 0.035499 (rank : 41) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K039, Q9CSR4, Q9D0W8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1143 homolog. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.035272 (rank : 42) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.034875 (rank : 43) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.034484 (rank : 44) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.034449 (rank : 45) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.032005 (rank : 46) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CP39A_MOUSE
|
||||||
| NC score | 0.031758 (rank : 47) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKJ9 | Gene names | Cyp39a1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (mCYP39A1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.031655 (rank : 48) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.031274 (rank : 49) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.030545 (rank : 50) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.030545 (rank : 51) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
MCES_HUMAN
|
||||||
| NC score | 0.029707 (rank : 52) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.025514 (rank : 53) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.025435 (rank : 54) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.025335 (rank : 55) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
NOL3_MOUSE
|
||||||
| NC score | 0.024495 (rank : 56) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein 3. | |||||
|
ERRFI_HUMAN
|
||||||
| NC score | 0.022483 (rank : 57) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
IRX2_HUMAN
|
||||||
| NC score | 0.022090 (rank : 58) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.021822 (rank : 59) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.021680 (rank : 60) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.021635 (rank : 61) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
DMXL2_HUMAN
|
||||||
| NC score | 0.020321 (rank : 62) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.019893 (rank : 63) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.019296 (rank : 64) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TNNT1_HUMAN
|
||||||
| NC score | 0.018728 (rank : 65) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P13805, O95472, Q16061 | Gene names | TNNT1, TNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, slow skeletal muscle (TnTs) (Slow skeletal muscle troponin T) (sTnT). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.018352 (rank : 66) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RBBP5_HUMAN
|
||||||
| NC score | 0.018254 (rank : 67) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
CP39A_HUMAN
|
||||||
| NC score | 0.017426 (rank : 68) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.017081 (rank : 69) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.016290 (rank : 70) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
VSX1_MOUSE
|
||||||
| NC score | 0.016143 (rank : 71) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.014692 (rank : 72) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
ASXL1_MOUSE
|
||||||
| NC score | 0.014305 (rank : 73) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.014272 (rank : 74) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.013458 (rank : 75) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
LRMP_HUMAN
|
||||||
| NC score | 0.013024 (rank : 76) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12912, Q8N301 | Gene names | LRMP, JAW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
TSH2_HUMAN
|
||||||
| NC score | 0.011938 (rank : 77) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.011784 (rank : 78) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.011289 (rank : 79) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.011069 (rank : 80) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.011033 (rank : 81) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.010878 (rank : 82) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PSMD2_MOUSE
|
||||||
| NC score | 0.010833 (rank : 83) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.010679 (rank : 84) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.009857 (rank : 85) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
ENPL_MOUSE
|
||||||
| NC score | 0.008516 (rank : 86) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08113, P11427 | Gene names | Hsp90b1, Tra-1, Tra1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (ERP99) (Polymorphic tumor rejection antigen 1) (Tumor rejection antigen gp96). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.008275 (rank : 87) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.008076 (rank : 88) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.008027 (rank : 89) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
ACK1_HUMAN
|
||||||
| NC score | 0.007120 (rank : 90) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.006603 (rank : 91) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
SH23A_HUMAN
|
||||||
| NC score | 0.006369 (rank : 92) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRG2, Q9Y2X4 | Gene names | SH2D3A, NSP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3A (Novel SH2-containing protein 1). | |||||
|
ACK1_MOUSE
|
||||||
| NC score | 0.006186 (rank : 93) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
PEG3_HUMAN
|
||||||
| NC score | 0.005873 (rank : 94) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9GZU2, P78418, Q5H9P9, Q7Z7H7, Q8TF75, Q9GZY2 | Gene names | PEG3, KIAA0287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein. | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.005486 (rank : 95) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.004840 (rank : 96) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.003793 (rank : 97) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.003606 (rank : 98) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.003425 (rank : 99) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.002811 (rank : 100) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||