Please be patient as the page loads
|
CLGN_MOUSE
|
||||||
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CALX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.966992 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CALX_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.968645 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CLGN_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.972935 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor. | |||||
|
CLGN_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
CALR_HUMAN
|
||||||
| θ value | 1.90147e-66 (rank : 5) | NC score | 0.877125 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
CALR_MOUSE
|
||||||
| θ value | 1.6097e-65 (rank : 6) | NC score | 0.879937 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14211 | Gene names | Calr | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60). | |||||
|
CALR3_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 7) | NC score | 0.815561 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L12, Q96LN3 | Gene names | CALR3, CRT2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR3_MOUSE
|
||||||
| θ value | 1.6813e-30 (rank : 8) | NC score | 0.805672 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.053528 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
SCG3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.075673 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P47867 | Gene names | Scg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretogranin-3 precursor (Secretogranin III) (SgIII). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.030929 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.022374 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.057783 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.057297 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NFL_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.020524 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
SPT6H_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.055649 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
CENPB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.051239 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.041595 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.022999 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.040274 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.038773 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
CCDC9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.056732 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.053850 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.044411 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
SPT6H_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.053442 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.009366 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CLC11_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.030006 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y240 | Gene names | CLEC11A, CLECSF3, LSLCL, SCGF | |||
|
Domain Architecture |
|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin) (p47) (C-type lectin superfamily member 3). | |||||
|
CN145_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.023037 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.012549 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.008971 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
RED_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.045627 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13123 | Gene names | IK, RED, RER | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.045439 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.023696 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.013986 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.017592 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.020425 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
K0690_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.025867 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JTH9, Q5JTH8, Q69YK4, Q96E87, Q9BUH3, Q9Y4C7 | Gene names | KIAA0690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0690. | |||||
|
AN32B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.016325 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.030785 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.043635 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.013708 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
DACH1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.019702 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
DPPA4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.027876 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CCG4, Q6P228, Q810Y6 | Gene names | Dppa4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmental pluripotency-associated protein 4. | |||||
|
SDCG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.032153 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
TNNT2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.026737 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.062821 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
FAM9A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.034101 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
MCES_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.032996 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
RED_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.041857 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1M8 | Gene names | Ik, Red, Rer | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
RRAGD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.016535 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQL2, Q7L8F9, Q9NPG0 | Gene names | RRAGD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related GTP-binding protein D (Rag D). | |||||
|
TNNT2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.027304 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
ABCF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.017495 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
ANGE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.031392 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNK9, O94859 | Gene names | ANGEL1, KIAA0759 | |||
|
Domain Architecture |
|
|||||
| Description | Angel homolog 1. | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.033918 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
DMN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.033144 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.046191 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.015779 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.020116 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NFL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.018692 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.016502 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.006479 (rank : 151) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.031272 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
AMPM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.025242 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08663 | Gene names | Metap2, Mnpep, P67eif2 | |||
|
Domain Architecture |

|
|||||
| Description | Methionine aminopeptidase 2 (EC 3.4.11.18) (MetAP 2) (Peptidase M 2) (Initiation factor 2-associated 67 kDa glycoprotein) (p67) (p67eIF2). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.026886 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.047375 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ICAL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.026433 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.046411 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NP1L3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.024611 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.020674 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.031852 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.023009 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.023831 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.017668 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.030970 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
BNC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.017456 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
CP39A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.025519 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKJ9 | Gene names | Cyp39a1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (mCYP39A1). | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.070563 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.055851 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.073079 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.021176 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PEG3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.002977 (rank : 154) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9GZU2, P78418, Q5H9P9, Q7Z7H7, Q8TF75, Q9GZY2 | Gene names | PEG3, KIAA0287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein. | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.014322 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.023111 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.017124 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.003032 (rank : 153) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.016198 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.007272 (rank : 145) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.010195 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
AMPM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.022703 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50579 | Gene names | METAP2, MNPEP, P67EIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Methionine aminopeptidase 2 (EC 3.4.11.18) (MetAP 2) (Peptidase M 2) (Initiation factor 2-associated 67 kDa glycoprotein) (p67) (p67eIF2). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.026992 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.017006 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
H6ST3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.043734 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
KI21B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.007395 (rank : 144) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
MED12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.017267 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.016651 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
NCA11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.004394 (rank : 152) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13591, Q15829, Q16180 | Gene names | NCAM1, NCAM | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 140 kDa isoform precursor (N-CAM 140) (NCAM-140) (CD56 antigen). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.024060 (rank : 78) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
PDXL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.015332 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.017607 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RRP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.024442 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.001633 (rank : 156) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.015764 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.017739 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.013703 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CD244_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.013654 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZW8, Q96T47, Q9NQD2, Q9NQD3, Q9Y288 | Gene names | CD244, 2B4 | |||
|
Domain Architecture |
|
|||||
| Description | Natural killer cell receptor 2B4 precursor (NKR2B4) (NK cell type I receptor protein 2B4) (h2B4) (CD244 antigen) (NK cell activation- inducing ligand) (NAIL). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.021020 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.007227 (rank : 147) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
HS90A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.009016 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
I28RA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.012853 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CGK5, Q6PEV1 | Gene names | Il28ra, Ifnlr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-28 receptor alpha chain precursor (IL-28R-alpha) (IL-28RA) (Cytokine receptor family 2 member 12) (Cytokine receptor class-II member 12) (CRF2-12) (Interferon lambda receptor 1) (IFN-lambda R1). | |||||
|
KI21B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.006990 (rank : 148) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
LMD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.014337 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.015680 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
NEK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.002530 (rank : 155) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.025294 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.022003 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.007261 (rank : 146) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.032611 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RBM13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.015493 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXY0, Q5U5T1, Q86UC4, Q96SY6 | Gene names | RBM13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAK16-like protein RBM13 (RNA-binding motif protein 13) (NNP78). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.006858 (rank : 149) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.019796 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.020734 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
STIM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.018881 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.033417 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.012946 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
ZN403_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.017752 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3C7, Q96T90, Q9GZR8, Q9H767 | Gene names | ZNF403, LCRG1, LZK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Laryngeal carcinoma-related protein 1). | |||||
|
ZN403_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.017635 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SV77, Q5SV75, Q5SV76, Q5SV78, Q6GVH6, Q6P9J4, Q920N4 | Gene names | Znf403, Ggnbp2, Zfp403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Gametogenetin-binding protein 2) (Dioxin-inducible factor 3) (DIF-3). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.008578 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.026253 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CD20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.009099 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11836, P08984, Q13963 | Gene names | MS4A1, CD20 | |||
|
Domain Architecture |
|
|||||
| Description | B-lymphocyte antigen CD20 (B-lymphocyte surface antigen B1) (Leu-16) (Bp35). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.023856 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.007980 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.014478 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
GSDM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.008251 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EST1 | Gene names | Gsdm1, Gsdm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin. | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.016230 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.016688 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
K0562_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.010792 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.029627 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.027482 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.007941 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.006634 (rank : 150) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.024231 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
SDCG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.023882 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |
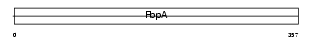
|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.057618 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.051918 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CC47_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.061684 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
CF010_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.064736 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.109087 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.055995 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.058200 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.069844 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.089403 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.101437 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.058487 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.067231 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.082076 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.060712 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CLGN_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
CLGN_HUMAN
|
||||||
| NC score | 0.972935 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor. | |||||
|
CALX_MOUSE
|
||||||
| NC score | 0.968645 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.966992 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CALR_MOUSE
|
||||||
| NC score | 0.879937 (rank : 5) | θ value | 1.6097e-65 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14211 | Gene names | Calr | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60). | |||||
|
CALR_HUMAN
|
||||||
| NC score | 0.877125 (rank : 6) | θ value | 1.90147e-66 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
CALR3_HUMAN
|
||||||
| NC score | 0.815561 (rank : 7) | θ value | 3.06405e-32 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L12, Q96LN3 | Gene names | CALR3, CRT2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR3_MOUSE
|
||||||
| NC score | 0.805672 (rank : 8) | θ value | 1.6813e-30 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.109087 (rank : 9) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.101437 (rank : 10) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.089403 (rank : 11) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.082076 (rank : 12) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SCG3_MOUSE
|
||||||
| NC score | 0.075673 (rank : 13) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P47867 | Gene names | Scg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretogranin-3 precursor (Secretogranin III) (SgIII). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.073079 (rank : 14) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.070563 (rank : 15) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.069844 (rank : 16) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.067231 (rank : 17) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.064736 (rank : 18) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.062821 (rank : 19) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.061684 (rank : 20) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.060712 (rank : 21) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.058487 (rank : 22) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.058200 (rank : 23) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.057783 (rank : 24) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.057618 (rank : 25) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.057297 (rank : 26) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
CCDC9_HUMAN
|
||||||
| NC score | 0.056732 (rank : 27) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.055995 (rank : 28) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.055851 (rank : 29) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SPT6H_HUMAN
|
||||||
| NC score | 0.055649 (rank : 30) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.053850 (rank : 31) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.053528 (rank : 32) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
SPT6H_MOUSE
|
||||||
| NC score | 0.053442 (rank : 33) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.051918 (rank : 34) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CENPB_HUMAN
|
||||||
| NC score | 0.051239 (rank : 35) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.047375 (rank : 36) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.046411 (rank : 37) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.046191 (rank : 38) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
RED_HUMAN
|
||||||
| NC score | 0.045627 (rank : 39) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13123 | Gene names | IK, RED, RER | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.045439 (rank : 40) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.044411 (rank : 41) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
H6ST3_MOUSE
|
||||||
| NC score | 0.043734 (rank : 42) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.043635 (rank : 43) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
RED_MOUSE
|
||||||
| NC score | 0.041857 (rank : 44) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1M8 | Gene names | Ik, Red, Rer | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.041595 (rank : 45) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.040274 (rank : 46) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.038773 (rank : 47) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
FAM9A_HUMAN
|
||||||
| NC score | 0.034101 (rank : 48) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.033918 (rank : 49) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.033417 (rank : 50) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
DMN_HUMAN
|
||||||
| NC score | 0.033144 (rank : 51) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
MCES_HUMAN
|
||||||
| NC score | 0.032996 (rank : 52) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.032611 (rank : 53) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
SDCG1_MOUSE
|
||||||
| NC score | 0.032153 (rank : 54) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.031852 (rank : 55) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
ANGE1_HUMAN
|
||||||
| NC score | 0.031392 (rank : 56) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNK9, O94859 | Gene names | ANGEL1, KIAA0759 | |||
|
Domain Architecture |

|
|||||
| Description | Angel homolog 1. | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.031272 (rank : 57) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.030970 (rank : 58) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.030929 (rank : 59) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.030785 (rank : 60) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CLC11_HUMAN
|
||||||
| NC score | 0.030006 (rank : 61) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y240 | Gene names | CLEC11A, CLECSF3, LSLCL, SCGF | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin) (p47) (C-type lectin superfamily member 3). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.029627 (rank : 62) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
DPPA4_MOUSE
|
||||||
| NC score | 0.027876 (rank : 63) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CCG4, Q6P228, Q810Y6 | Gene names | Dppa4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmental pluripotency-associated protein 4. | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.027482 (rank : 64) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
TNNT2_MOUSE
|
||||||
| NC score | 0.027304 (rank : 65) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.026992 (rank : 66) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.026886 (rank : 67) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
TNNT2_HUMAN
|
||||||
| NC score | 0.026737 (rank : 68) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.026433 (rank : 69) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.026253 (rank : 70) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
K0690_HUMAN
|
||||||
| NC score | 0.025867 (rank : 71) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JTH9, Q5JTH8, Q69YK4, Q96E87, Q9BUH3, Q9Y4C7 | Gene names | KIAA0690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0690. | |||||
|
CP39A_MOUSE
|
||||||
| NC score | 0.025519 (rank : 72) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKJ9 | Gene names | Cyp39a1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (mCYP39A1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.025294 (rank : 73) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AMPM2_MOUSE
|
||||||
| NC score | 0.025242 (rank : 74) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08663 | Gene names | Metap2, Mnpep, P67eif2 | |||
|
Domain Architecture |

|
|||||
| Description | Methionine aminopeptidase 2 (EC 3.4.11.18) (MetAP 2) (Peptidase M 2) (Initiation factor 2-associated 67 kDa glycoprotein) (p67) (p67eIF2). | |||||
|
NP1L3_HUMAN
|
||||||
| NC score | 0.024611 (rank : 75) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.024442 (rank : 76) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.024231 (rank : 77) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.024060 (rank : 78) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
SDCG1_HUMAN
|
||||||
| NC score | 0.023882 (rank : 79) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.023856 (rank : 80) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.023831 (rank : 81) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.023696 (rank : 82) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.023111 (rank : 83) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.023037 (rank : 84) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.023009 (rank : 85) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.022999 (rank : 86) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
AMPM2_HUMAN
|
||||||
| NC score | 0.022703 (rank : 87) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50579 | Gene names | METAP2, MNPEP, P67EIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Methionine aminopeptidase 2 (EC 3.4.11.18) (MetAP 2) (Peptidase M 2) (Initiation factor 2-associated 67 kDa glycoprotein) (p67) (p67eIF2). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.022374 (rank : 88) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.022003 (rank : 89) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.021176 (rank : 90) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.021020 (rank : 91) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.020734 (rank : 92) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.020674 (rank : 93) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.020524 (rank : 94) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.020425 (rank : 95) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.020116 (rank : 96) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.019796 (rank : 97) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
DACH1_HUMAN
|
||||||
| NC score | 0.019702 (rank : 98) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
STIM1_MOUSE
|
||||||
| NC score | 0.018881 (rank : 99) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
NFL_HUMAN
|
||||||
| NC score | 0.018692 (rank : 100) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
ZN403_HUMAN
|
||||||
| NC score | 0.017752 (rank : 101) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3C7, Q96T90, Q9GZR8, Q9H767 | Gene names | ZNF403, LCRG1, LZK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Laryngeal carcinoma-related protein 1). | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.017739 (rank : 102) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.017668 (rank : 103) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
ZN403_MOUSE
|
||||||
| NC score | 0.017635 (rank : 104) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SV77, Q5SV75, Q5SV76, Q5SV78, Q6GVH6, Q6P9J4, Q920N4 | Gene names | Znf403, Ggnbp2, Zfp403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Gametogenetin-binding protein 2) (Dioxin-inducible factor 3) (DIF-3). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.017607 (rank : 105) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.017592 (rank : 106) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ABCF1_HUMAN
|
||||||
| NC score | 0.017495 (rank : 107) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
BNC2_MOUSE
|
||||||
| NC score | 0.017456 (rank : 108) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
MED12_HUMAN
|
||||||
| NC score | 0.017267 (rank : 109) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.017124 (rank : 110) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.017006 (rank : 111) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.016688 (rank : 112) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.016651 (rank : 113) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
RRAGD_HUMAN
|
||||||
| NC score | 0.016535 (rank : 114) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQL2, Q7L8F9, Q9NPG0 | Gene names | RRAGD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related GTP-binding protein D (Rag D). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.016502 (rank : 115) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
AN32B_HUMAN
|
||||||
| NC score | 0.016325 (rank : 116) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.016230 (rank : 117) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.016198 (rank : 118) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.015779 (rank : 119) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.015764 (rank : 120) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.015680 (rank : 121) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
RBM13_HUMAN
|
||||||
| NC score | 0.015493 (rank : 122) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXY0, Q5U5T1, Q86UC4, Q96SY6 | Gene names | RBM13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAK16-like protein RBM13 (RNA-binding motif protein 13) (NNP78). | |||||
|
PDXL2_HUMAN
|
||||||
| NC score | 0.015332 (rank : 123) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.014478 (rank : 124) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
LMD1_HUMAN
|
||||||
| NC score | 0.014337 (rank : 125) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.014322 (rank : 126) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.013986 (rank : 127) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.013708 (rank : 128) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.013703 (rank : 129) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CD244_HUMAN
|
||||||
| NC score | 0.013654 (rank : 130) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZW8, Q96T47, Q9NQD2, Q9NQD3, Q9Y288 | Gene names | CD244, 2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cell receptor 2B4 precursor (NKR2B4) (NK cell type I receptor protein 2B4) (h2B4) (CD244 antigen) (NK cell activation- inducing ligand) (NAIL). | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.012946 (rank : 131) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
I28RA_MOUSE
|
||||||
| NC score | 0.012853 (rank : 132) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CGK5, Q6PEV1 | Gene names | Il28ra, Ifnlr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-28 receptor alpha chain precursor (IL-28R-alpha) (IL-28RA) (Cytokine receptor family 2 member 12) (Cytokine receptor class-II member 12) (CRF2-12) (Interferon lambda receptor 1) (IFN-lambda R1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.012549 (rank : 133) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
K0562_MOUSE
|
||||||
| NC score | 0.010792 (rank : 134) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.010195 (rank : 135) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.009366 (rank : 136) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CD20_HUMAN
|
||||||
| NC score | 0.009099 (rank : 137) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11836, P08984, Q13963 | Gene names | MS4A1, CD20 | |||
|
Domain Architecture |

|
|||||
| Description | B-lymphocyte antigen CD20 (B-lymphocyte surface antigen B1) (Leu-16) (Bp35). | |||||
|
HS90A_HUMAN
|
||||||
| NC score | 0.009016 (rank : 138) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.008971 (rank : 139) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.008578 (rank : 140) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
GSDM1_MOUSE
|
||||||
| NC score | 0.008251 (rank : 141) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EST1 | Gene names | Gsdm1, Gsdm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin. | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.007980 (rank : 142) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.007941 (rank : 143) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
KI21B_MOUSE
|
||||||
| NC score | 0.007395 (rank : 144) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.007272 (rank : 145) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.007261 (rank : 146) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.007227 (rank : 147) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
KI21B_HUMAN
|
||||||
| NC score | 0.006990 (rank : 148) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.006858 (rank : 149) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.006634 (rank : 150) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.006479 (rank : 151) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
NCA11_HUMAN
|
||||||
| NC score | 0.004394 (rank : 152) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13591, Q15829, Q16180 | Gene names | NCAM1, NCAM | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 140 kDa isoform precursor (N-CAM 140) (NCAM-140) (CD56 antigen). | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.003032 (rank : 153) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
PEG3_HUMAN
|
||||||
| NC score | 0.002977 (rank : 154) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9GZU2, P78418, Q5H9P9, Q7Z7H7, Q8TF75, Q9GZY2 | Gene names | PEG3, KIAA0287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein. | |||||
|
NEK1_HUMAN
|
||||||
| NC score | 0.002530 (rank : 155) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.001633 (rank : 156) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||