Please be patient as the page loads
|
CALX_MOUSE
|
||||||
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CALX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991095 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CALX_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CLGN_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.973101 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor. | |||||
|
CLGN_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.968645 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
CALR_HUMAN
|
||||||
| θ value | 6.76295e-64 (rank : 5) | NC score | 0.896161 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
CALR_MOUSE
|
||||||
| θ value | 2.56992e-63 (rank : 6) | NC score | 0.896774 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P14211 | Gene names | Calr | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60). | |||||
|
CALR3_HUMAN
|
||||||
| θ value | 2.67802e-36 (rank : 7) | NC score | 0.829469 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L12, Q96LN3 | Gene names | CALR3, CRT2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR3_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 8) | NC score | 0.819090 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.094576 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.064925 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.055573 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.058851 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.085797 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
VSX1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.018302 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.057981 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.071716 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.066415 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
H6ST3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.068210 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.010443 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.064106 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.050252 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.068448 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.063416 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CP39A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.033032 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKJ9 | Gene names | Cyp39a1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (mCYP39A1). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.084417 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.034459 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.031400 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.032090 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.020715 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.008569 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.054390 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
ANK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.009624 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.016709 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.013084 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.023736 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.016383 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
ADA30_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.007946 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.025581 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.032868 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.098893 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
IRX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.021545 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.046356 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.026944 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.023635 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
ZFY1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | -0.000466 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
LMNB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.003335 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
OSTP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.035018 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10451, Q15681, Q15682, Q15683, Q8NBK2, Q96IZ1 | Gene names | SPP1, OPN | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Urinary stone protein) (Nephropontin) (Uropontin). | |||||
|
PDE3B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.013736 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
RFTN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.014559 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.026212 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.011816 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CCD94_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.015668 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BW85, O75270, Q9H862, Q9NW16 | Gene names | CCDC94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 94. | |||||
|
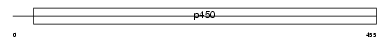
CP39A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.018666 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
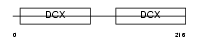
DCDC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.014465 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |
|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.006036 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.020715 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.029854 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TOP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.031657 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
ACK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.002654 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
AN32A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.022954 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.022954 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.005978 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
ERCC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.012103 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.024371 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.008223 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
TF2AY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.014967 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
TNNT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.020319 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13805, O95472, Q16061 | Gene names | TNNT1, TNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, slow skeletal muscle (TnTs) (Slow skeletal muscle troponin T) (sTnT). | |||||
|
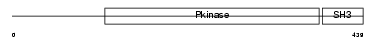
ACK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.001844 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |
|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.019529 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
GTF2I_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.011194 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.017339 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.013229 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NPHP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.007239 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
PLOD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.005723 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0E2 | Gene names | Plod1, Plod | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 precursor (EC 1.14.11.4) (Lysyl hydroxylase 1) (LH1). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.019345 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.014478 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.061209 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.055979 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
CALX_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.991095 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CLGN_HUMAN
|
||||||
| NC score | 0.973101 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor. | |||||
|
CLGN_MOUSE
|
||||||
| NC score | 0.968645 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
CALR_MOUSE
|
||||||
| NC score | 0.896774 (rank : 5) | θ value | 2.56992e-63 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P14211 | Gene names | Calr | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60). | |||||
|
CALR_HUMAN
|
||||||
| NC score | 0.896161 (rank : 6) | θ value | 6.76295e-64 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
CALR3_HUMAN
|
||||||
| NC score | 0.829469 (rank : 7) | θ value | 2.67802e-36 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L12, Q96LN3 | Gene names | CALR3, CRT2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR3_MOUSE
|
||||||
| NC score | 0.819090 (rank : 8) | θ value | 5.22648e-32 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.098893 (rank : 9) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.094576 (rank : 10) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.085797 (rank : 11) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.084417 (rank : 12) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.071716 (rank : 13) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.068448 (rank : 14) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
H6ST3_MOUSE
|
||||||
| NC score | 0.068210 (rank : 15) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.066415 (rank : 16) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.064925 (rank : 17) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.064106 (rank : 18) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.063416 (rank : 19) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.061209 (rank : 20) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.058851 (rank : 21) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.057981 (rank : 22) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.055979 (rank : 23) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.055573 (rank : 24) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.054390 (rank : 25) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.050252 (rank : 26) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.046356 (rank : 27) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
OSTP_HUMAN
|
||||||
| NC score | 0.035018 (rank : 28) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10451, Q15681, Q15682, Q15683, Q8NBK2, Q96IZ1 | Gene names | SPP1, OPN | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Urinary stone protein) (Nephropontin) (Uropontin). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.034459 (rank : 29) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
CP39A_MOUSE
|
||||||
| NC score | 0.033032 (rank : 30) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKJ9 | Gene names | Cyp39a1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (mCYP39A1). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.032868 (rank : 31) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.032090 (rank : 32) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
TOP1_MOUSE
|
||||||
| NC score | 0.031657 (rank : 33) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.031400 (rank : 34) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.029854 (rank : 35) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.026944 (rank : 36) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.026212 (rank : 37) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.025581 (rank : 38) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.024371 (rank : 39) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.023736 (rank : 40) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.023635 (rank : 41) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
AN32A_MOUSE
|
||||||
| NC score | 0.022954 (rank : 42) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| NC score | 0.022954 (rank : 43) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
IRX2_HUMAN
|
||||||
| NC score | 0.021545 (rank : 44) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.020715 (rank : 45) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.020715 (rank : 46) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TNNT1_HUMAN
|
||||||
| NC score | 0.020319 (rank : 47) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13805, O95472, Q16061 | Gene names | TNNT1, TNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, slow skeletal muscle (TnTs) (Slow skeletal muscle troponin T) (sTnT). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.019529 (rank : 48) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.019345 (rank : 49) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
CP39A_HUMAN
|
||||||
| NC score | 0.018666 (rank : 50) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
VSX1_MOUSE
|
||||||
| NC score | 0.018302 (rank : 51) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.017339 (rank : 52) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.016709 (rank : 53) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.016383 (rank : 54) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
CCD94_HUMAN
|
||||||
| NC score | 0.015668 (rank : 55) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BW85, O75270, Q9H862, Q9NW16 | Gene names | CCDC94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 94. | |||||
|
TF2AY_HUMAN
|
||||||
| NC score | 0.014967 (rank : 56) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
RFTN1_HUMAN
|
||||||
| NC score | 0.014559 (rank : 57) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.014478 (rank : 58) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
DCDC2_HUMAN
|
||||||
| NC score | 0.014465 (rank : 59) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |

|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
PDE3B_HUMAN
|
||||||
| NC score | 0.013736 (rank : 60) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.013229 (rank : 61) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.013084 (rank : 62) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
ERCC4_MOUSE
|
||||||
| NC score | 0.012103 (rank : 63) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.011816 (rank : 64) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
GTF2I_HUMAN
|
||||||
| NC score | 0.011194 (rank : 65) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.010443 (rank : 66) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.009624 (rank : 67) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.008569 (rank : 68) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.008223 (rank : 69) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
ADA30_HUMAN
|
||||||
| NC score | 0.007946 (rank : 70) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
NPHP3_MOUSE
|
||||||
| NC score | 0.007239 (rank : 71) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.006036 (rank : 72) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.005978 (rank : 73) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
PLOD1_MOUSE
|
||||||
| NC score | 0.005723 (rank : 74) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0E2 | Gene names | Plod1, Plod | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 precursor (EC 1.14.11.4) (Lysyl hydroxylase 1) (LH1). | |||||
|
LMNB2_HUMAN
|
||||||
| NC score | 0.003335 (rank : 75) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
ACK1_HUMAN
|
||||||
| NC score | 0.002654 (rank : 76) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
ACK1_MOUSE
|
||||||
| NC score | 0.001844 (rank : 77) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
ZFY1_MOUSE
|
||||||
| NC score | -0.000466 (rank : 78) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
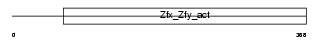
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||