Please be patient as the page loads
|
GTF2I_HUMAN
|
||||||
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GTF2I_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 151 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
GTF2I_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979415 (rank : 2) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
GT2D1_HUMAN
|
||||||
| θ value | 2.71407e-105 (rank : 3) | NC score | 0.865123 (rank : 3) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHL9, O95444, Q75MX7, Q86UM3, Q8WVC4, Q9UHK8, Q9UI91 | Gene names | GTF2IRD1, CREAM1, GTF3, MUSTRD1, RBAP2, WBSCR11, WBSCR12 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Muscle TFII-I repeat domain-containing protein 1) (General transcription factor III) (Slow- muscle-fiber enhancer-binding protein) (USE B1-binding protein) (MusTRD1/BEN) (Williams-Beuren syndrome chromosome region 11 protein). | |||||
|
GT2D1_MOUSE
|
||||||
| θ value | 1.95407e-87 (rank : 4) | NC score | 0.845763 (rank : 4) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 5) | NC score | 0.024632 (rank : 72) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 6) | NC score | 0.024916 (rank : 71) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.085431 (rank : 6) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 8) | NC score | 0.082873 (rank : 9) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.058836 (rank : 15) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.085063 (rank : 7) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.071986 (rank : 12) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.084146 (rank : 8) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
MORC3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.048747 (rank : 21) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14149, Q9UEZ2 | Gene names | MORC3, KIAA0136, ZCWCC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 3 (Zinc finger CW-type coiled- coil domain protein 3). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.048986 (rank : 20) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.038710 (rank : 31) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
ESX1L_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.022755 (rank : 81) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.034784 (rank : 42) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ASPH_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.072197 (rank : 11) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.066622 (rank : 14) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.089304 (rank : 5) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.045028 (rank : 26) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.033424 (rank : 51) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
NTKL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.045581 (rank : 25) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EQC5, Q3TST7, Q3UKU0, Q8K222 | Gene names | Scyl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like protein (SCY1-like protein 1) (105 kDa kinase- like protein) (Mitosis-associated kinase-like protein NTKL). | |||||
|
HS105_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.037456 (rank : 34) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.043932 (rank : 27) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.003040 (rank : 147) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.021774 (rank : 91) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.033521 (rank : 48) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.018541 (rank : 107) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.035630 (rank : 38) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.042919 (rank : 28) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.028133 (rank : 63) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
AMELY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.035721 (rank : 37) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
CCD38_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.034275 (rank : 44) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
CJ047_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.035031 (rank : 41) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.012558 (rank : 125) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.081965 (rank : 10) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.038338 (rank : 32) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYST2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.051805 (rank : 18) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95251 | Gene names | MYST2, HBO1, HBOa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST2 (EC 2.3.1.48) (MYST protein 2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (Histone acetyltransferase binding to ORC1). | |||||
|
MYST2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.051993 (rank : 17) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SVQ0, Q5SVQ1, Q5SVQ2, Q5SVQ3, Q5SVQ7, Q6PGC6, Q80Y65 | Gene names | Myst2, Hbo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST2 (EC 2.3.1.48) (MYST protein 2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (Histone acetyltransferase binding to ORC1). | |||||
|
NXPH2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.022248 (rank : 85) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61199, Q8BIN8 | Gene names | Nxph2, Nph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexophilin-2 precursor. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.028205 (rank : 62) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RPA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.023573 (rank : 77) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.008028 (rank : 138) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ZFY1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.003663 (rank : 146) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.047083 (rank : 23) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
HS105_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.023883 (rank : 74) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
JIP3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.025417 (rank : 68) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.035276 (rank : 40) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.034343 (rank : 43) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SSX3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.025044 (rank : 70) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99909, O60223, Q5JQZ3 | Gene names | SSX3 | |||
|
Domain Architecture |
|
|||||
| Description | Protein SSX3. | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.011458 (rank : 128) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ZFY19_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.026447 (rank : 66) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.034250 (rank : 45) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.032568 (rank : 53) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.033520 (rank : 49) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.037478 (rank : 33) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.029371 (rank : 58) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.027129 (rank : 64) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.022058 (rank : 87) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.014275 (rank : 122) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.018744 (rank : 105) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.020395 (rank : 96) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.006340 (rank : 141) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.022036 (rank : 88) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.025321 (rank : 69) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
PP1R7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.019249 (rank : 102) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UM45, Q9CV31, Q9Z105 | Gene names | Ppp1r7, Sds22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 7 (Protein phosphatase 1 regulatory subunit 22). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.046706 (rank : 24) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.035918 (rank : 36) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.004660 (rank : 144) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.067443 (rank : 13) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
ANTR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.023735 (rank : 76) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.018969 (rank : 104) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CD68_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.022266 (rank : 84) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P34810, Q96BI7 | Gene names | CD68 | |||
|
Domain Architecture |
|
|||||
| Description | Macrosialin precursor (GP110) (CD68 antigen). | |||||
|
IF3X_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.032667 (rank : 52) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.011339 (rank : 130) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.047359 (rank : 22) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.049291 (rank : 19) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NCKX4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.017089 (rank : 111) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFF2, Q8N8U6, Q8NCX1, Q8NFF0, Q8NFF1 | Gene names | SLC24A4, NCKX4 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 4 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 4) (Solute carrier family 24 member 4). | |||||
|
NOP14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.024060 (rank : 73) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.030167 (rank : 56) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.023220 (rank : 79) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.021776 (rank : 90) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.023882 (rank : 75) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.032217 (rank : 54) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ZCH14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.030288 (rank : 55) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ANTR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.022068 (rank : 86) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |
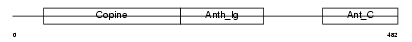
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.010603 (rank : 133) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CLCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.025577 (rank : 67) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09496 | Gene names | CLTA | |||
|
Domain Architecture |
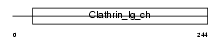
|
|||||
| Description | Clathrin light chain A (Lca). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.030165 (rank : 57) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CQ032_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.034099 (rank : 46) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SYH2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf32 homolog. | |||||
|
CRTAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.019043 (rank : 103) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75718 | Gene names | CRTAP, CASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.010736 (rank : 132) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
IF39_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.020237 (rank : 97) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
JHD2C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.019960 (rank : 101) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
MKL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.016399 (rank : 112) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
NCKX4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.016216 (rank : 114) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CGQ8, Q8BLL5, Q8BLL7 | Gene names | Slc24a4, Nckx4 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 4 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 4). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.026550 (rank : 65) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NOL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.033497 (rank : 50) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |
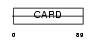
|
|||||
| Description | Nucleolar protein 3. | |||||
|
NTKL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.035611 (rank : 39) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96KG9, Q96G50, Q96KG8, Q96KH1, Q9HAW5, Q9HBL3, Q9NR53 | Gene names | SCYL1, GKLP, NTKL, TAPK, TEIF, TRAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like protein (SCY1-like protein 1) (Teratoma- associated tyrosine kinase) (Telomerase transcriptional element- interacting factor) (Telomerase regulation-associated protein). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.014581 (rank : 121) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.004110 (rank : 145) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.021286 (rank : 92) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.020081 (rank : 99) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.018087 (rank : 108) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.033828 (rank : 47) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
ZFY2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.002428 (rank : 149) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
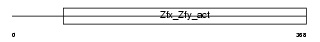
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.016112 (rank : 115) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.013583 (rank : 123) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CTCF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.000822 (rank : 151) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.013343 (rank : 124) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
KLH17_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.002836 (rank : 148) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6TDP3 | Gene names | Klhl17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 17 (Actinfilin). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.028396 (rank : 61) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MCM3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.009428 (rank : 135) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.038817 (rank : 30) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MOT8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.011791 (rank : 126) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
PHAR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.017296 (rank : 110) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0D0, Q3MJ93, Q5JSJ2 | Gene names | PHACTR1, KIAA1733, RPEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatase and actin regulator 1. | |||||
|
PHAR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.017393 (rank : 109) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q2M3X8, Q80VL9, Q8C873 | Gene names | Phactr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 1. | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.016228 (rank : 113) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.022561 (rank : 83) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SMN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.029285 (rank : 59) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |
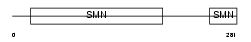
|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
SNAI_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.001072 (rank : 150) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95863, Q9P113, Q9UBP7, Q9UHH7 | Gene names | SNAI1, SNAH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein SNAI1 (Protein snail homolog) (Protein sna). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.020181 (rank : 98) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SPT6H_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.014744 (rank : 120) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
SYK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.020679 (rank : 94) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MN1 | Gene names | Kars | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl-tRNA synthetase (EC 6.1.1.6) (Lysine--tRNA ligase) (LysRS). | |||||
|
TCAL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.019981 (rank : 100) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921P9 | Gene names | Tceal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1). | |||||
|
U5S1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.011496 (rank : 127) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08810 | Gene names | Eftud2, Snrp116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.022846 (rank : 80) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.009450 (rank : 134) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AHI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.004953 (rank : 143) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.008967 (rank : 136) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
CALX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.011194 (rank : 131) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.014990 (rank : 117) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.020549 (rank : 95) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.023265 (rank : 78) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.018693 (rank : 106) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
FYB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.014968 (rank : 118) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.022006 (rank : 89) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
KLH11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.005992 (rank : 142) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVR0 | Gene names | KLHL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.008935 (rank : 137) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.039313 (rank : 29) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.020853 (rank : 93) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NPFF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.000323 (rank : 152) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5X5, Q96RV1, Q9NR49 | Gene names | NPFFR2, GPR74, NPFF2, NPGPR | |||
|
Domain Architecture |

|
|||||
| Description | Neuropeptide FF receptor 2 (Neuropeptide G-protein coupled receptor) (G-protein coupled receptor 74) (G-protein coupled receptor HLWAR77). | |||||
|
RNH2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.015290 (rank : 116) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TBB1, Q6PK48, Q9HAF7 | Gene names | RNASEH2B, DLEU8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease H2 subunit B (RNase H2 subunit B) (Ribonuclease HI subunit B) (Aicardi-Goutieres syndrome 2 protein) (AGS2) (Deleted in lymphocytic leukemia 8). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.037293 (rank : 35) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.006496 (rank : 140) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.022649 (rank : 82) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TF3C1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.011364 (rank : 129) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.028875 (rank : 60) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.014927 (rank : 119) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.007779 (rank : 139) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.055036 (rank : 16) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
GTF2I_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 151 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
GTF2I_MOUSE
|
||||||
| NC score | 0.979415 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
GT2D1_HUMAN
|
||||||
| NC score | 0.865123 (rank : 3) | θ value | 2.71407e-105 (rank : 3) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHL9, O95444, Q75MX7, Q86UM3, Q8WVC4, Q9UHK8, Q9UI91 | Gene names | GTF2IRD1, CREAM1, GTF3, MUSTRD1, RBAP2, WBSCR11, WBSCR12 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Muscle TFII-I repeat domain-containing protein 1) (General transcription factor III) (Slow- muscle-fiber enhancer-binding protein) (USE B1-binding protein) (MusTRD1/BEN) (Williams-Beuren syndrome chromosome region 11 protein). | |||||
|
GT2D1_MOUSE
|
||||||
| NC score | 0.845763 (rank : 4) | θ value | 1.95407e-87 (rank : 4) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.089304 (rank : 5) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.085431 (rank : 6) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.085063 (rank : 7) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.084146 (rank : 8) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.082873 (rank : 9) | θ value | 0.0193708 (rank : 8) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.081965 (rank : 10) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ASPH_HUMAN
|
||||||
| NC score | 0.072197 (rank : 11) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.071986 (rank : 12) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.067443 (rank : 13) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.066622 (rank : 14) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.058836 (rank : 15) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.055036 (rank : 16) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MYST2_MOUSE
|
||||||
| NC score | 0.051993 (rank : 17) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SVQ0, Q5SVQ1, Q5SVQ2, Q5SVQ3, Q5SVQ7, Q6PGC6, Q80Y65 | Gene names | Myst2, Hbo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST2 (EC 2.3.1.48) (MYST protein 2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (Histone acetyltransferase binding to ORC1). | |||||
|
MYST2_HUMAN
|
||||||
| NC score | 0.051805 (rank : 18) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95251 | Gene names | MYST2, HBO1, HBOa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST2 (EC 2.3.1.48) (MYST protein 2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (Histone acetyltransferase binding to ORC1). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.049291 (rank : 19) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.048986 (rank : 20) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
MORC3_HUMAN
|
||||||
| NC score | 0.048747 (rank : 21) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14149, Q9UEZ2 | Gene names | MORC3, KIAA0136, ZCWCC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 3 (Zinc finger CW-type coiled- coil domain protein 3). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.047359 (rank : 22) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.047083 (rank : 23) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.046706 (rank : 24) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
NTKL_MOUSE
|
||||||
| NC score | 0.045581 (rank : 25) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EQC5, Q3TST7, Q3UKU0, Q8K222 | Gene names | Scyl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like protein (SCY1-like protein 1) (105 kDa kinase- like protein) (Mitosis-associated kinase-like protein NTKL). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.045028 (rank : 26) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.043932 (rank : 27) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.042919 (rank : 28) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.039313 (rank : 29) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.038817 (rank : 30) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.038710 (rank : 31) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.038338 (rank : 32) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.037478 (rank : 33) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.037456 (rank : 34) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.037293 (rank : 35) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.035918 (rank : 36) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
AMELY_HUMAN
|
||||||
| NC score | 0.035721 (rank : 37) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.035630 (rank : 38) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NTKL_HUMAN
|
||||||
| NC score | 0.035611 (rank : 39) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96KG9, Q96G50, Q96KG8, Q96KH1, Q9HAW5, Q9HBL3, Q9NR53 | Gene names | SCYL1, GKLP, NTKL, TAPK, TEIF, TRAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like protein (SCY1-like protein 1) (Teratoma- associated tyrosine kinase) (Telomerase transcriptional element- interacting factor) (Telomerase regulation-associated protein). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.035276 (rank : 40) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
CJ047_HUMAN
|
||||||
| NC score | 0.035031 (rank : 41) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.034784 (rank : 42) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.034343 (rank : 43) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
CCD38_HUMAN
|
||||||
| NC score | 0.034275 (rank : 44) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.034250 (rank : 45) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CQ032_MOUSE
|
||||||
| NC score | 0.034099 (rank : 46) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SYH2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf32 homolog. | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.033828 (rank : 47) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.033521 (rank : 48) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.033520 (rank : 49) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
NOL3_MOUSE
|
||||||
| NC score | 0.033497 (rank : 50) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein 3. | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.033424 (rank : 51) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
IF3X_HUMAN
|
||||||
| NC score | 0.032667 (rank : 52) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.032568 (rank : 53) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.032217 (rank : 54) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ZCH14_HUMAN
|
||||||
| NC score | 0.030288 (rank : 55) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.030167 (rank : 56) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.030165 (rank : 57) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.029371 (rank : 58) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SMN_HUMAN
|
||||||
| NC score | 0.029285 (rank : 59) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |

|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.028875 (rank : 60) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.028396 (rank : 61) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.028205 (rank : 62) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.028133 (rank : 63) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.027129 (rank : 64) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.026550 (rank : 65) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZFY19_HUMAN
|
||||||
| NC score | 0.026447 (rank : 66) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
CLCA_HUMAN
|
||||||
| NC score | 0.025577 (rank : 67) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09496 | Gene names | CLTA | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain A (Lca). | |||||
|
JIP3_MOUSE
|
||||||
| NC score | 0.025417 (rank : 68) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.025321 (rank : 69) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
SSX3_HUMAN
|
||||||
| NC score | 0.025044 (rank : 70) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99909, O60223, Q5JQZ3 | Gene names | SSX3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SSX3. | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.024916 (rank : 71) | θ value | 0.00509761 (rank : 6) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.024632 (rank : 72) | θ value | 0.000461057 (rank : 5) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
NOP14_MOUSE
|
||||||
| NC score | 0.024060 (rank : 73) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
HS105_MOUSE
|
||||||
| NC score | 0.023883 (rank : 74) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.023882 (rank : 75) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
ANTR1_HUMAN
|
||||||
| NC score | 0.023735 (rank : 76) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
RPA1_MOUSE
|
||||||
| NC score | 0.023573 (rank : 77) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.023265 (rank : 78) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.023220 (rank : 79) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.022846 (rank : 80) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
ESX1L_HUMAN
|
||||||
| NC score | 0.022755 (rank : 81) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.022649 (rank : 82) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.022561 (rank : 83) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CD68_HUMAN
|
||||||
| NC score | 0.022266 (rank : 84) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P34810, Q96BI7 | Gene names | CD68 | |||
|
Domain Architecture |

|
|||||
| Description | Macrosialin precursor (GP110) (CD68 antigen). | |||||
|
NXPH2_MOUSE
|
||||||
| NC score | 0.022248 (rank : 85) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61199, Q8BIN8 | Gene names | Nxph2, Nph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexophilin-2 precursor. | |||||
|
ANTR1_MOUSE
|
||||||
| NC score | 0.022068 (rank : 86) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.022058 (rank : 87) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.022036 (rank : 88) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.022006 (rank : 89) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.021776 (rank : 90) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.021774 (rank : 91) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.021286 (rank : 92) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.020853 (rank : 93) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
SYK_MOUSE
|
||||||
| NC score | 0.020679 (rank : 94) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MN1 | Gene names | Kars | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl-tRNA synthetase (EC 6.1.1.6) (Lysine--tRNA ligase) (LysRS). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.020549 (rank : 95) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.020395 (rank : 96) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.020237 (rank : 97) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.020181 (rank : 98) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.020081 (rank : 99) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TCAL1_MOUSE
|
||||||
| NC score | 0.019981 (rank : 100) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921P9 | Gene names | Tceal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1). | |||||
|
JHD2C_HUMAN
|
||||||
| NC score | 0.019960 (rank : 101) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
PP1R7_MOUSE
|
||||||
| NC score | 0.019249 (rank : 102) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UM45, Q9CV31, Q9Z105 | Gene names | Ppp1r7, Sds22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 7 (Protein phosphatase 1 regulatory subunit 22). | |||||
|
CRTAP_HUMAN
|
||||||
| NC score | 0.019043 (rank : 103) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75718 | Gene names | CRTAP, CASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.018969 (rank : 104) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.018744 (rank : 105) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.018693 (rank : 106) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.018541 (rank : 107) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.018087 (rank : 108) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
PHAR1_MOUSE
|
||||||
| NC score | 0.017393 (rank : 109) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q2M3X8, Q80VL9, Q8C873 | Gene names | Phactr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 1. | |||||
|
PHAR1_HUMAN
|
||||||
| NC score | 0.017296 (rank : 110) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0D0, Q3MJ93, Q5JSJ2 | Gene names | PHACTR1, KIAA1733, RPEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatase and actin regulator 1. | |||||
|
NCKX4_HUMAN
|
||||||
| NC score | 0.017089 (rank : 111) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFF2, Q8N8U6, Q8NCX1, Q8NFF0, Q8NFF1 | Gene names | SLC24A4, NCKX4 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 4 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 4) (Solute carrier family 24 member 4). | |||||
|
MKL2_HUMAN
|
||||||
| NC score | 0.016399 (rank : 112) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.016228 (rank : 113) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
NCKX4_MOUSE
|
||||||
| NC score | 0.016216 (rank : 114) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CGQ8, Q8BLL5, Q8BLL7 | Gene names | Slc24a4, Nckx4 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 4 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 4). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.016112 (rank : 115) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
RNH2B_HUMAN
|
||||||
| NC score | 0.015290 (rank : 116) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TBB1, Q6PK48, Q9HAF7 | Gene names | RNASEH2B, DLEU8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease H2 subunit B (RNase H2 subunit B) (Ribonuclease HI subunit B) (Aicardi-Goutieres syndrome 2 protein) (AGS2) (Deleted in lymphocytic leukemia 8). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.014990 (rank : 117) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.014968 (rank : 118) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.014927 (rank : 119) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
SPT6H_HUMAN
|
||||||
| NC score | 0.014744 (rank : 120) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.014581 (rank : 121) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.014275 (rank : 122) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.013583 (rank : 123) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.013343 (rank : 124) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.012558 (rank : 125) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
MOT8_HUMAN
|
||||||
| NC score | 0.011791 (rank : 126) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
U5S1_MOUSE
|
||||||
| NC score | 0.011496 (rank : 127) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08810 | Gene names | Eftud2, Snrp116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.011458 (rank : 128) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
TF3C1_HUMAN
|
||||||
| NC score | 0.011364 (rank : 129) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.011339 (rank : 130) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
CALX_MOUSE
|
||||||
| NC score | 0.011194 (rank : 131) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.010736 (rank : 132) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.010603 (rank : 133) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.009450 (rank : 134) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
MCM3_HUMAN
|
||||||
| NC score | 0.009428 (rank : 135) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.008967 (rank : 136) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.008935 (rank : 137) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.008028 (rank : 138) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.007779 (rank : 139) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.006496 (rank : 140) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.006340 (rank : 141) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
KLH11_HUMAN
|
||||||
| NC score | 0.005992 (rank : 142) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVR0 | Gene names | KLHL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
AHI1_HUMAN
|
||||||
| NC score | 0.004953 (rank : 143) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.004660 (rank : 144) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.004110 (rank : 145) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
ZFY1_MOUSE
|
||||||
| NC score | 0.003663 (rank : 146) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.003040 (rank : 147) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
KLH17_MOUSE
|
||||||
| NC score | 0.002836 (rank : 148) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6TDP3 | Gene names | Klhl17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 17 (Actinfilin). | |||||
|
ZFY2_MOUSE
|
||||||
| NC score | 0.002428 (rank : 149) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
SNAI_HUMAN
|
||||||
| NC score | 0.001072 (rank : 150) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95863, Q9P113, Q9UBP7, Q9UHH7 | Gene names | SNAI1, SNAH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein SNAI1 (Protein snail homolog) (Protein sna). | |||||
|
CTCF_MOUSE
|
||||||
| NC score | 0.000822 (rank : 151) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
NPFF2_HUMAN
|
||||||
| NC score | 0.000323 (rank : 152) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5X5, Q96RV1, Q9NR49 | Gene names | NPFFR2, GPR74, NPFF2, NPGPR | |||
|
Domain Architecture |

|
|||||
| Description | Neuropeptide FF receptor 2 (Neuropeptide G-protein coupled receptor) (G-protein coupled receptor 74) (G-protein coupled receptor HLWAR77). | |||||