Please be patient as the page loads
|
MYST2_HUMAN
|
||||||
| SwissProt Accessions | O95251 | Gene names | MYST2, HBO1, HBOa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST2 (EC 2.3.1.48) (MYST protein 2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (Histone acetyltransferase binding to ORC1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MYST2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O95251 | Gene names | MYST2, HBO1, HBOa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST2 (EC 2.3.1.48) (MYST protein 2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (Histone acetyltransferase binding to ORC1). | |||||
|
MYST2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999803 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q5SVQ0, Q5SVQ1, Q5SVQ2, Q5SVQ3, Q5SVQ7, Q6PGC6, Q80Y65 | Gene names | Myst2, Hbo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST2 (EC 2.3.1.48) (MYST protein 2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (Histone acetyltransferase binding to ORC1). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 4.05562e-101 (rank : 3) | NC score | 0.732619 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 6.91785e-101 (rank : 4) | NC score | 0.718720 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 1.70393e-99 (rank : 5) | NC score | 0.766949 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 4.95767e-99 (rank : 6) | NC score | 0.728469 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST1_HUMAN
|
||||||
| θ value | 3.00772e-96 (rank : 7) | NC score | 0.941328 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H7Z6, Q659G0, Q7LC17, Q8IY59, Q8WYB4, Q8WZ14, Q9HAC5, Q9NR35 | Gene names | MYST1, MOF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable histone acetyltransferase MYST1 (EC 2.3.1.48) (MYST protein 1) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 1) (hMOF). | |||||
|
MYST1_MOUSE
|
||||||
| θ value | 3.00772e-96 (rank : 8) | NC score | 0.940873 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D1P2, Q3UIY0, Q8BJ69, Q8BJ76, Q8CI73 | Gene names | Myst1, Mof | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable histone acetyltransferase MYST1 (EC 2.3.1.48) (MYST protein 1) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 1). | |||||
|
TIP60_HUMAN
|
||||||
| θ value | 1.95407e-87 (rank : 9) | NC score | 0.927248 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
TIP60_MOUSE
|
||||||
| θ value | 1.95407e-87 (rank : 10) | NC score | 0.927035 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 11) | NC score | 0.236282 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 12) | NC score | 0.228700 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
LMBTL_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 13) | NC score | 0.141134 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 14) | NC score | 0.220524 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 15) | NC score | 0.222305 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
ST18_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 16) | NC score | 0.210654 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
ST18_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 17) | NC score | 0.210422 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
OFD1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 18) | NC score | 0.049366 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
TNIP3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.046752 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
ILDR1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.049906 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86SU0, Q6ZP61, Q7Z578 | Gene names | ILDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin-like domain-containing receptor 1 precursor. | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.045947 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.030634 (rank : 99) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.036005 (rank : 96) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.023667 (rank : 117) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
PRDM5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.002990 (rank : 149) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQX1 | Gene names | PRDM5, PFM2 | |||
|
Domain Architecture |
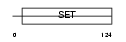
|
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
PRDM5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.003165 (rank : 148) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CXE0 | Gene names | Prdm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.034385 (rank : 97) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RP9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.053513 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TA86 | Gene names | RP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinitis pigmentosa 9 protein (Pim-1-associated protein) (PAP-1). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.028182 (rank : 102) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
GTF2I_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.039799 (rank : 95) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.025030 (rank : 113) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.026365 (rank : 108) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.024916 (rank : 114) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
AT10A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.013103 (rank : 138) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
GTF2I_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.051805 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.022648 (rank : 121) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.005982 (rank : 146) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.027005 (rank : 105) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
TAK1L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.049235 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P57077 | Gene names | TAK1L, C21orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TAK1-like protein. | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.019922 (rank : 128) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
REST_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.024761 (rank : 115) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RU17_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.025413 (rank : 112) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.011674 (rank : 139) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.022264 (rank : 122) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
RU17_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.024074 (rank : 116) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
FIP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.027174 (rank : 104) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.021508 (rank : 124) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.009215 (rank : 143) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.028026 (rank : 103) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.030510 (rank : 100) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ABRA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.016467 (rank : 130) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUZ1, Q3UU01, Q8BLH3, Q8K431 | Gene names | Abra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.006081 (rank : 145) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.026271 (rank : 109) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.028832 (rank : 101) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.001397 (rank : 151) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.016338 (rank : 131) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.118887 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.064052 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.022956 (rank : 120) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.014278 (rank : 135) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.023625 (rank : 118) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.007642 (rank : 144) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.010836 (rank : 140) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.009757 (rank : 142) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.045959 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.021058 (rank : 126) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.021058 (rank : 127) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.022974 (rank : 119) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.025656 (rank : 111) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.026184 (rank : 110) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.044909 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.033378 (rank : 98) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.015449 (rank : 133) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DREB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.021570 (rank : 123) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
F107B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.026673 (rank : 106) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H098 | Gene names | FAM107B, C10orf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM107B. | |||||
|
F107B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.026637 (rank : 107) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3TGF2 | Gene names | Fam107b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM107B. | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.042610 (rank : 94) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
TOP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.013875 (rank : 137) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
TRI56_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.010749 (rank : 141) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
ZBT38_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.002519 (rank : 150) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.048536 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.016153 (rank : 132) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
DREB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.016835 (rank : 129) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
IF33_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.014082 (rank : 136) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WK2 | Gene names | Eif3s3 | |||
|
Domain Architecture |
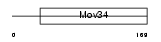
|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | -0.000039 (rank : 152) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
REST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.021233 (rank : 125) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.014813 (rank : 134) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
VRK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.003810 (rank : 147) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99986 | Gene names | VRK1 | |||
|
Domain Architecture |
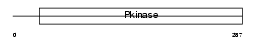
|
|||||
| Description | Serine/threonine-protein kinase VRK1 (EC 2.7.11.1) (Vaccinia-related kinase 1). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.050857 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.092428 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.091373 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.071020 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.057250 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
ASPH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.070446 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.079936 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.082001 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.080287 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.078253 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.081089 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CJ049_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.063074 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WVF2 | Gene names | C10orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf49 precursor. | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.198386 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.173185 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.078646 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58269 | Gene names | Dpf3, Cerd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-finger protein Dpf3 (cer-d4). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.090904 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
HRX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.062512 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.083740 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.077755 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.059198 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.067511 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.066736 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.061780 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.059251 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.082122 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.085791 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051983 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
LSP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.056107 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.092917 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.080115 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.085638 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.066433 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.078969 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NEST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.052438 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.059706 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.080951 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.050569 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.057574 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.110693 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.109484 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.087625 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.088057 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.222613 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.222073 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.102870 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.103192 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.059268 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.060836 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PHF7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.061613 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.178238 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.178514 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.085397 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.063436 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.050938 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.051170 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.062631 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.050738 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.066336 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.057194 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.076458 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.068174 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.060802 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.056762 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050211 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
MYST2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O95251 | Gene names | MYST2, HBO1, HBOa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST2 (EC 2.3.1.48) (MYST protein 2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (Histone acetyltransferase binding to ORC1). | |||||
|
MYST2_MOUSE
|
||||||
| NC score | 0.999803 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q5SVQ0, Q5SVQ1, Q5SVQ2, Q5SVQ3, Q5SVQ7, Q6PGC6, Q80Y65 | Gene names | Myst2, Hbo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST2 (EC 2.3.1.48) (MYST protein 2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (Histone acetyltransferase binding to ORC1). | |||||
|
MYST1_HUMAN
|
||||||
| NC score | 0.941328 (rank : 3) | θ value | 3.00772e-96 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H7Z6, Q659G0, Q7LC17, Q8IY59, Q8WYB4, Q8WZ14, Q9HAC5, Q9NR35 | Gene names | MYST1, MOF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable histone acetyltransferase MYST1 (EC 2.3.1.48) (MYST protein 1) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 1) (hMOF). | |||||
|
MYST1_MOUSE
|
||||||
| NC score | 0.940873 (rank : 4) | θ value | 3.00772e-96 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D1P2, Q3UIY0, Q8BJ69, Q8BJ76, Q8CI73 | Gene names | Myst1, Mof | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable histone acetyltransferase MYST1 (EC 2.3.1.48) (MYST protein 1) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 1). | |||||
|
TIP60_HUMAN
|
||||||
| NC score | 0.927248 (rank : 5) | θ value | 1.95407e-87 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
TIP60_MOUSE
|
||||||
| NC score | 0.927035 (rank : 6) | θ value | 1.95407e-87 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.766949 (rank : 7) | θ value | 1.70393e-99 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.732619 (rank : 8) | θ value | 4.05562e-101 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.728469 (rank : 9) | θ value | 4.95767e-99 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.718720 (rank : 10) | θ value | 6.91785e-101 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.236282 (rank : 11) | θ value | 1.06045e-08 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.228700 (rank : 12) | θ value | 1.38499e-08 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.222613 (rank : 13) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.222305 (rank : 14) | θ value | 3.77169e-06 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.222073 (rank : 15) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.220524 (rank : 16) | θ value | 3.77169e-06 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
ST18_HUMAN
|
||||||
| NC score | 0.210654 (rank : 17) | θ value | 3.19293e-05 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
ST18_MOUSE
|
||||||
| NC score | 0.210422 (rank : 18) | θ value | 3.19293e-05 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.198386 (rank : 19) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.178514 (rank : 20) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.178238 (rank : 21) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.173185 (rank : 22) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
LMBTL_HUMAN
|
||||||
| NC score | 0.141134 (rank : 23) | θ value | 4.45536e-07 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.118887 (rank : 24) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.110693 (rank : 25) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.109484 (rank : 26) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.103192 (rank : 27) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.102870 (rank : 28) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.092917 (rank : 29) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.092428 (rank : 30) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.091373 (rank : 31) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.090904 (rank : 32) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.088057 (rank : 33) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.087625 (rank : 34) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.085791 (rank : 35) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.085638 (rank : 36) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.085397 (rank : 37) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.083740 (rank : 38) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.082122 (rank : 39) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.082001 (rank : 40) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.081089 (rank : 41) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.080951 (rank : 42) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.080287 (rank : 43) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.080115 (rank : 44) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.079936 (rank : 45) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.078969 (rank : 46) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
DPF3_MOUSE
|
||||||
| NC score | 0.078646 (rank : 47) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58269 | Gene names | Dpf3, Cerd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-finger protein Dpf3 (cer-d4). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.078253 (rank : 48) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.077755 (rank : 49) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.076458 (rank : 50) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.071020 (rank : 51) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ASPH_HUMAN
|
||||||
| NC score | 0.070446 (rank : 52) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.068174 (rank : 53) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.067511 (rank : 54) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.066736 (rank : 55) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.066433 (rank : 56) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.066336 (rank : 57) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.064052 (rank : 58) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.063436 (rank : 59) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
CJ049_HUMAN
|
||||||
| NC score | 0.063074 (rank : 60) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WVF2 | Gene names | C10orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf49 precursor. | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.062631 (rank : 61) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.062512 (rank : 62) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.061780 (rank : 63) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
PHF7_HUMAN
|
||||||
| NC score | 0.061613 (rank : 64) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.060836 (rank : 65) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.060802 (rank : 66) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.059706 (rank : 67) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.059268 (rank : 68) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
JAD1D_MOUSE
|
||||||
| NC score | 0.059251 (rank : 69) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.059198 (rank : 70) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.057574 (rank : 71) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.057250 (rank : 72) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.057194 (rank : 73) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.056762 (rank : 74) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
LSP1_HUMAN
|
||||||
| NC score | 0.056107 (rank : 75) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
RP9_HUMAN
|
||||||
| NC score | 0.053513 (rank : 76) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TA86 | Gene names | RP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinitis pigmentosa 9 protein (Pim-1-associated protein) (PAP-1). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.052438 (rank : 77) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.051983 (rank : 78) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
GTF2I_HUMAN
|
||||||
| NC score | 0.051805 (rank : 79) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.051170 (rank : 80) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.050938 (rank : 81) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.050857 (rank : 82) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.050738 (rank : 83) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.050569 (rank : 84) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.050211 (rank : 85) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ILDR1_HUMAN
|
||||||
| NC score | 0.049906 (rank : 86) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86SU0, Q6ZP61, Q7Z578 | Gene names | ILDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin-like domain-containing receptor 1 precursor. | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.049366 (rank : 87) | θ value | 0.0193708 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
TAK1L_HUMAN
|
||||||
| NC score | 0.049235 (rank : 88) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P57077 | Gene names | TAK1L, C21orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TAK1-like protein. | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.048536 (rank : 89) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
TNIP3_HUMAN
|
||||||
| NC score | 0.046752 (rank : 90) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.045959 (rank : 91) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.045947 (rank : 92) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.044909 (rank : 93) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.042610 (rank : 94) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
GTF2I_MOUSE
|
||||||
| NC score | 0.039799 (rank : 95) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.036005 (rank : 96) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.034385 (rank : 97) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.033378 (rank : 98) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.030634 (rank : 99) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.030510 (rank : 100) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.028832 (rank : 101) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.028182 (rank : 102) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.028026 (rank : 103) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
FIP1_MOUSE
|
||||||
| NC score | 0.027174 (rank : 104) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.027005 (rank : 105) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
F107B_HUMAN
|
||||||
| NC score | 0.026673 (rank : 106) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H098 | Gene names | FAM107B, C10orf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM107B. | |||||
|
F107B_MOUSE
|
||||||
| NC score | 0.026637 (rank : 107) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3TGF2 | Gene names | Fam107b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM107B. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.026365 (rank : 108) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.026271 (rank : 109) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.026184 (rank : 110) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.025656 (rank : 111) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
RU17_HUMAN
|
||||||
| NC score | 0.025413 (rank : 112) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.025030 (rank : 113) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.024916 (rank : 114) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.024761 (rank : 115) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RU17_MOUSE
|
||||||
| NC score | 0.024074 (rank : 116) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.023667 (rank : 117) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.023625 (rank : 118) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.022974 (rank : 119) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.022956 (rank : 120) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.022648 (rank : 121) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.022264 (rank : 122) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
DREB_HUMAN
|
||||||
| NC score | 0.021570 (rank : 123) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.021508 (rank : 124) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.021233 (rank : 125) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.021058 (rank : 126) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.021058 (rank : 127) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.019922 (rank : 128) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
DREB_MOUSE
|
||||||
| NC score | 0.016835 (rank : 129) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
ABRA_MOUSE
|
||||||
| NC score | 0.016467 (rank : 130) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUZ1, Q3UU01, Q8BLH3, Q8K431 | Gene names | Abra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.016338 (rank : 131) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.016153 (rank : 132) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.015449 (rank : 133) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.014813 (rank : 134) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.014278 (rank : 135) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
IF33_MOUSE
|
||||||
| NC score | 0.014082 (rank : 136) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WK2 | Gene names | Eif3s3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
TOP1_HUMAN
|
||||||
| NC score | 0.013875 (rank : 137) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
AT10A_HUMAN
|
||||||
| NC score | 0.013103 (rank : 138) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.011674 (rank : 139) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.010836 (rank : 140) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
TRI56_HUMAN
|
||||||
| NC score | 0.010749 (rank : 141) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.009757 (rank : 142) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.009215 (rank : 143) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.007642 (rank : 144) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.006081 (rank : 145) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.005982 (rank : 146) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
VRK1_HUMAN
|
||||||
| NC score | 0.003810 (rank : 147) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99986 | Gene names | VRK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase VRK1 (EC 2.7.11.1) (Vaccinia-related kinase 1). | |||||
|
PRDM5_MOUSE
|
||||||
| NC score | 0.003165 (rank : 148) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CXE0 | Gene names | Prdm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
PRDM5_HUMAN
|
||||||
| NC score | 0.002990 (rank : 149) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQX1 | Gene names | PRDM5, PFM2 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
ZBT38_HUMAN
|
||||||
| NC score | 0.002519 (rank : 150) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.001397 (rank : 151) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | -0.000039 (rank : 152) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||