Please be patient as the page loads
|
CLCA_HUMAN
|
||||||
| SwissProt Accessions | P09496 | Gene names | CLTA | |||
|
Domain Architecture |
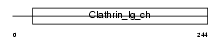
|
|||||
| Description | Clathrin light chain A (Lca). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CLCA_HUMAN
|
||||||
| θ value | 6.44497e-115 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P09496 | Gene names | CLTA | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain A (Lca). | |||||
|
CLCA_MOUSE
|
||||||
| θ value | 3.79596e-99 (rank : 2) | NC score | 0.982110 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O08585 | Gene names | Clta | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain A (Lca). | |||||
|
CLCB_HUMAN
|
||||||
| θ value | 7.71989e-68 (rank : 3) | NC score | 0.933750 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
CLCB_MOUSE
|
||||||
| θ value | 8.53533e-67 (rank : 4) | NC score | 0.933705 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 5) | NC score | 0.186829 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 6) | NC score | 0.155990 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.108706 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.114936 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.108358 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.075748 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.061493 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.142375 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.105322 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
NXF2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.054506 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZY0, Q9BXU4, Q9NSS1, Q9NX66 | Gene names | NXF2, TAPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear RNA export factor 2 (TAP-like protein 2) (TAPL-2). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.082822 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.076535 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
PERI_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.034471 (rank : 128) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P41219, Q8N577 | Gene names | PRPH | |||
|
Domain Architecture |
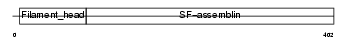
|
|||||
| Description | Peripherin. | |||||
|
PERI_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.033629 (rank : 130) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15331, O35688, O35689 | Gene names | Prph, Prph1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
INCE_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.072312 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.066110 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.065431 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.071995 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
PEX5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.033658 (rank : 129) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
RN168_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.062228 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.077619 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.071595 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.061839 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.063020 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
KI13A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.025963 (rank : 136) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
KI13A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.028338 (rank : 135) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.061149 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.022220 (rank : 148) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.031072 (rank : 133) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.013543 (rank : 159) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
CCD19_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.055143 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.070546 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
INCE_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.082094 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.065835 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
RN168_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.051738 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80XJ2 | Gene names | Rnf168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.096295 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.047004 (rank : 119) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MYO6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.022843 (rank : 144) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.063221 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
SIN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.053793 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BPZ7, Q00426 | Gene names | MAPKAP1, SIN1 | |||
|
Domain Architecture |
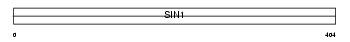
|
|||||
| Description | Stress-activated map kinase-interacting protein 1 (SAPK-interacting protein 1) (Putative Ras inhibitor JC310). | |||||
|
U2AFL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.123111 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.063992 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.093471 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.074050 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.074362 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
K1H5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.020461 (rank : 152) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92764, O76012, Q92651 | Gene names | KRTHA5, HHA5, HKA5 | |||
|
Domain Architecture |
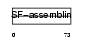
|
|||||
| Description | Keratin, type I cuticular Ha5 (Hair keratin, type I Ha5). | |||||
|
LRC59_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.021714 (rank : 149) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q922Q8, Q3TJ35, Q3TLC7, Q3TWT9, Q3TX86 | Gene names | Lrrc59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.076176 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
TRI35_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.022827 (rank : 145) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 465 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8C006, Q6ZPY0, Q7TQL7, Q810V7, Q8BVY9, Q8VID4, Q9CW74, Q9D208 | Gene names | Trim35, Hls5, Kiaa1098, Mair | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 35 (Macrophage-derived apoptosis- inducing RBCC protein) (Protein MAIR) (Protein Nc8) (Hemopoietic lineage switch protein 5). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.035769 (rank : 126) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.074590 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.024260 (rank : 140) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
GBP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.032650 (rank : 132) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96PP9, Q86T99 | Gene names | GBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate-binding protein 4 (GTP-binding protein 4) (Guanine nucleotide-binding protein 4) (GBP-4). | |||||
|
GTF2I_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.025577 (rank : 138) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
GTF2I_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.025560 (rank : 139) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
K1C16_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.020196 (rank : 153) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z2K1 | Gene names | Krt16, Krt1-16 | |||
|
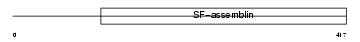
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.076652 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.035562 (rank : 127) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
NRL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.017657 (rank : 155) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
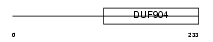
Domain Architecture |
|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.064460 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.047539 (rank : 118) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.014972 (rank : 157) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.025749 (rank : 137) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.022953 (rank : 142) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
GCP60_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.038083 (rank : 124) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
K0408_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.028542 (rank : 134) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZU52, O43158, Q5TF20, Q7L2M2 | Gene names | KIAA0408 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0408. | |||||
|
LRC59_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.019382 (rank : 154) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.043343 (rank : 121) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.047973 (rank : 117) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.023096 (rank : 141) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.022768 (rank : 146) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.014140 (rank : 158) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
OTU6B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.020576 (rank : 151) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N6M0, Q9NTA4, Q9Y387 | Gene names | OTUD6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 6B. | |||||
|
PACE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.022596 (rank : 147) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBQ7, Q6NY01, Q8BQC9, Q8BRJ1 | Gene names | Scyl3, Pace1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-associating with the carboxyl-terminal domain of ezrin (Ezrin- binding protein PACE-1) (SCY1-like protein 3). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.042038 (rank : 123) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
ARHG4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.005137 (rank : 161) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.044907 (rank : 120) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GLE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.094587 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
K2C6E_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.022867 (rank : 143) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.048781 (rank : 116) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
SARDH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.010460 (rank : 160) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LB7 | Gene names | Sardh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.021456 (rank : 150) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SMC4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.036363 (rank : 125) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.014976 (rank : 156) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
TXLNG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.033397 (rank : 131) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.042494 (rank : 122) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.050365 (rank : 111) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.061046 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CCD21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.054579 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.050205 (rank : 114) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.056438 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.054373 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCD45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.053132 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96GE4, Q96M81 | Gene names | CCDC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.065071 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.061546 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CCD91_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.060404 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
CDC37_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.053984 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
CF060_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.056928 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CI039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.050237 (rank : 112) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CI093_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053422 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CING_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050476 (rank : 109) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.064906 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.050233 (rank : 113) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.058247 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.050598 (rank : 106) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.053392 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.055658 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.055022 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
DESP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.050397 (rank : 110) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.050526 (rank : 108) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
GAS8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.052485 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
GEMI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.052160 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
GOG8A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050646 (rank : 105) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.057248 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.061599 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.061698 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.054304 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.055719 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.053728 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.051755 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.051254 (rank : 98) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.053643 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.057123 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
LRC45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.052528 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.055333 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.051132 (rank : 99) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
MERL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.069233 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.069119 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051919 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.051321 (rank : 97) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.051417 (rank : 95) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.051359 (rank : 96) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
NDE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.051935 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
NEMO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.055271 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
NEST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.054663 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NOP14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.053446 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.057999 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.050186 (rank : 115) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.053515 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.050796 (rank : 101) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.053688 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.050738 (rank : 103) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.052803 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.054386 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051775 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
SH24A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.053805 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.050686 (rank : 104) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.051566 (rank : 94) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.051639 (rank : 93) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.059566 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.050831 (rank : 100) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.050769 (rank : 102) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.050583 (rank : 107) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
U2AF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.064380 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01081 | Gene names | U2AF1, U2AF35 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
U2AF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.064360 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D883, Q564E4, Q99LX2, Q9CZ98 | Gene names | U2af1 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
WDR67_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.055749 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
ZWINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.058504 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
CLCA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.44497e-115 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P09496 | Gene names | CLTA | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain A (Lca). | |||||
|
CLCA_MOUSE
|
||||||
| NC score | 0.982110 (rank : 2) | θ value | 3.79596e-99 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O08585 | Gene names | Clta | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain A (Lca). | |||||
|
CLCB_HUMAN
|
||||||
| NC score | 0.933750 (rank : 3) | θ value | 7.71989e-68 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
CLCB_MOUSE
|
||||||
| NC score | 0.933705 (rank : 4) | θ value | 8.53533e-67 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.186829 (rank : 5) | θ value | 0.0148317 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.155990 (rank : 6) | θ value | 0.0148317 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.142375 (rank : 7) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
U2AFL_MOUSE
|
||||||
| NC score | 0.123111 (rank : 8) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.114936 (rank : 9) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.108706 (rank : 10) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.108358 (rank : 11) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.105322 (rank : 12) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.096295 (rank : 13) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
GLE1_HUMAN
|
||||||
| NC score | 0.094587 (rank : 14) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.093471 (rank : 15) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.082822 (rank : 16) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.082094 (rank : 17) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.077619 (rank : 18) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.076652 (rank : 19) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.076535 (rank : 20) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.076176 (rank : 21) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.075748 (rank : 22) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.074590 (rank : 23) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.074362 (rank : 24) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.074050 (rank : 25) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.072312 (rank : 26) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.071995 (rank : 27) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.071595 (rank : 28) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.070546 (rank : 29) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.069233 (rank : 30) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.069119 (rank : 31) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.066110 (rank : 32) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.065835 (rank : 33) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.065431 (rank : 34) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.065071 (rank : 35) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.064906 (rank : 36) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.064460 (rank : 37) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
U2AF1_HUMAN
|
||||||
| NC score | 0.064380 (rank : 38) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01081 | Gene names | U2AF1, U2AF35 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
U2AF1_MOUSE
|
||||||
| NC score | 0.064360 (rank : 39) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D883, Q564E4, Q99LX2, Q9CZ98 | Gene names | U2af1 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.063992 (rank : 40) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.063221 (rank : 41) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.063020 (rank : 42) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
RN168_HUMAN
|
||||||
| NC score | 0.062228 (rank : 43) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.061839 (rank : 44) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.061698 (rank : 45) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.061599 (rank : 46) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.061546 (rank : 47) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.061493 (rank : 48) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.061149 (rank : 49) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.061046 (rank : 50) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CCD91_MOUSE
|
||||||
| NC score | 0.060404 (rank : 51) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.059566 (rank : 52) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
ZWINT_MOUSE
|
||||||
| NC score | 0.058504 (rank : 53) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.058247 (rank : 54) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.057999 (rank : 55) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.057248 (rank : 56) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.057123 (rank : 57) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.056928 (rank : 58) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.056438 (rank : 59) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
WDR67_HUMAN
|
||||||
| NC score | 0.055749 (rank : 60) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.055719 (rank : 61) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.055658 (rank : 62) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.055333 (rank : 63) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NEMO_HUMAN
|
||||||
| NC score | 0.055271 (rank : 64) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
CCD19_HUMAN
|
||||||
| NC score | 0.055143 (rank : 65) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.055022 (rank : 66) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.054663 (rank : 67) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.054579 (rank : 68) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
NXF2_HUMAN
|
||||||
| NC score | 0.054506 (rank : 69) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZY0, Q9BXU4, Q9NSS1, Q9NX66 | Gene names | NXF2, TAPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear RNA export factor 2 (TAP-like protein 2) (TAPL-2). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.054386 (rank : 70) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.054373 (rank : 71) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.054304 (rank : 72) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
CDC37_HUMAN
|
||||||
| NC score | 0.053984 (rank : 73) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
SH24A_HUMAN
|
||||||
| NC score | 0.053805 (rank : 74) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
SIN1_HUMAN
|
||||||
| NC score | 0.053793 (rank : 75) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BPZ7, Q00426 | Gene names | MAPKAP1, SIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-activated map kinase-interacting protein 1 (SAPK-interacting protein 1) (Putative Ras inhibitor JC310). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.053728 (rank : 76) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.053688 (rank : 77) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.053643 (rank : 78) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.053515 (rank : 79) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.053446 (rank : 80) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.053422 (rank : 81) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.053392 (rank : 82) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CCD45_HUMAN
|
||||||
| NC score | 0.053132 (rank : 83) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96GE4, Q96M81 | Gene names | CCDC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.052803 (rank : 84) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.052528 (rank : 85) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.052485 (rank : 86) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
GEMI_MOUSE
|
||||||
| NC score | 0.052160 (rank : 87) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.051935 (rank : 88) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.051919 (rank : 89) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.051775 (rank : 90) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.051755 (rank : 91) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
RN168_MOUSE
|
||||||
| NC score | 0.051738 (rank : 92) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80XJ2 | Gene names | Rnf168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.051639 (rank : 93) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.051566 (rank : 94) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.051417 (rank : 95) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.051359 (rank : 96) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.051321 (rank : 97) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.051254 (rank : 98) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.051132 (rank : 99) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.050831 (rank : 100) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.050796 (rank : 101) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.050769 (rank : 102) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.050738 (rank : 103) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.050686 (rank : 104) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
GOG8A_HUMAN
|
||||||
| NC score | 0.050646 (rank : 105) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.050598 (rank : 106) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.050583 (rank : 107) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.050526 (rank : 108) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.050476 (rank : 109) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.050397 (rank : 110) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.050365 (rank : 111) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.050237 (rank : 112) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.050233 (rank : 113) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.050205 (rank : 114) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.050186 (rank : 115) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.048781 (rank : 116) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.047973 (rank : 117) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.047539 (rank : 118) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.047004 (rank : 119) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.044907 (rank : 120) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.043343 (rank : 121) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.042494 (rank : 122) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.042038 (rank : 123) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
GCP60_HUMAN
|
||||||
| NC score | 0.038083 (rank : 124) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.036363 (rank : 125) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.035769 (rank : 126) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.035562 (rank : 127) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PERI_HUMAN
|
||||||
| NC score | 0.034471 (rank : 128) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P41219, Q8N577 | Gene names | PRPH | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
PEX5_HUMAN
|
||||||
| NC score | 0.033658 (rank : 129) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
PERI_MOUSE
|
||||||
| NC score | 0.033629 (rank : 130) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15331, O35688, O35689 | Gene names | Prph, Prph1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
TXLNG_HUMAN
|
||||||
| NC score | 0.033397 (rank : 131) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
GBP4_HUMAN
|
||||||
| NC score | 0.032650 (rank : 132) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96PP9, Q86T99 | Gene names | GBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate-binding protein 4 (GTP-binding protein 4) (Guanine nucleotide-binding protein 4) (GBP-4). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.031072 (rank : 133) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
K0408_HUMAN
|
||||||
| NC score | 0.028542 (rank : 134) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZU52, O43158, Q5TF20, Q7L2M2 | Gene names | KIAA0408 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0408. | |||||
|
KI13A_MOUSE
|
||||||
| NC score | 0.028338 (rank : 135) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
KI13A_HUMAN
|
||||||
| NC score | 0.025963 (rank : 136) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.025749 (rank : 137) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
GTF2I_HUMAN
|
||||||
| NC score | 0.025577 (rank : 138) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
GTF2I_MOUSE
|
||||||
| NC score | 0.025560 (rank : 139) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.024260 (rank : 140) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.023096 (rank : 141) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.022953 (rank : 142) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
K2C6E_HUMAN
|
||||||
| NC score | 0.022867 (rank : 143) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
MYO6_MOUSE
|
||||||
| NC score | 0.022843 (rank : 144) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
TRI35_MOUSE
|
||||||
| NC score | 0.022827 (rank : 145) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 465 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8C006, Q6ZPY0, Q7TQL7, Q810V7, Q8BVY9, Q8VID4, Q9CW74, Q9D208 | Gene names | Trim35, Hls5, Kiaa1098, Mair | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 35 (Macrophage-derived apoptosis- inducing RBCC protein) (Protein MAIR) (Protein Nc8) (Hemopoietic lineage switch protein 5). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.022768 (rank : 146) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
PACE1_MOUSE
|
||||||
| NC score | 0.022596 (rank : 147) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBQ7, Q6NY01, Q8BQC9, Q8BRJ1 | Gene names | Scyl3, Pace1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-associating with the carboxyl-terminal domain of ezrin (Ezrin- binding protein PACE-1) (SCY1-like protein 3). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.022220 (rank : 148) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
LRC59_MOUSE
|
||||||
| NC score | 0.021714 (rank : 149) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q922Q8, Q3TJ35, Q3TLC7, Q3TWT9, Q3TX86 | Gene names | Lrrc59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.021456 (rank : 150) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
OTU6B_HUMAN
|
||||||
| NC score | 0.020576 (rank : 151) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N6M0, Q9NTA4, Q9Y387 | Gene names | OTUD6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 6B. | |||||
|
K1H5_HUMAN
|
||||||
| NC score | 0.020461 (rank : 152) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92764, O76012, Q92651 | Gene names | KRTHA5, HHA5, HKA5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha5 (Hair keratin, type I Ha5). | |||||
|
K1C16_MOUSE
|
||||||
| NC score | 0.020196 (rank : 153) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z2K1 | Gene names | Krt16, Krt1-16 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
LRC59_HUMAN
|
||||||
| NC score | 0.019382 (rank : 154) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
NRL_HUMAN
|
||||||
| NC score | 0.017657 (rank : 155) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
Domain Architecture |

|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.014976 (rank : 156) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.014972 (rank : 157) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.014140 (rank : 158) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.013543 (rank : 159) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
SARDH_MOUSE
|
||||||
| NC score | 0.010460 (rank : 160) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LB7 | Gene names | Sardh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH). | |||||
|
ARHG4_MOUSE
|
||||||
| NC score | 0.005137 (rank : 161) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||