Please be patient as the page loads
|
U2AF1_HUMAN
|
||||||
| SwissProt Accessions | Q01081 | Gene names | U2AF1, U2AF35 | |||
|
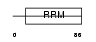
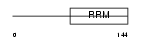
Domain Architecture |
|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
U2AF1_HUMAN
|
||||||
| θ value | 1.44581e-90 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01081 | Gene names | U2AF1, U2AF35 | |||
|
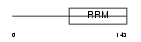
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
U2AF1_MOUSE
|
||||||
| θ value | 1.44581e-90 (rank : 2) | NC score | 0.999707 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D883, Q564E4, Q99LX2, Q9CZ98 | Gene names | U2af1 | |||
|
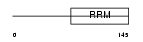
Domain Architecture |
|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 3) | NC score | 0.692101 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 4) | NC score | 0.659222 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 5) | NC score | 0.665524 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
U2AFL_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 6) | NC score | 0.730026 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
SPF45_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 7) | NC score | 0.212708 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 8) | NC score | 0.212067 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
PABP1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.120431 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.117431 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.126380 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
PABP3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.117097 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H361, Q8NHV0, Q9H086 | Gene names | PABPC3, PABP3, PABPL3 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 3 (Poly(A)-binding protein 3) (PABP 3) (Testis-specific poly(A)-binding protein). | |||||
|
PABP5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.117978 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96DU9, Q6P529, Q9UFE5 | Gene names | PABPC5, PABP5 | |||
|
Domain Architecture |
|
|||||
| Description | Polyadenylate-binding protein 5 (Poly(A)-binding protein 5) (PABP 5). | |||||
|
RBM4B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.111938 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.111906 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.112223 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.112001 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.117579 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.039165 (rank : 86) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MYEF2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.096352 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2K5, Q6NUM5, Q7L388, Q7Z4B7, Q9H922, Q9NUQ1 | Gene names | MYEF2, KIAA1341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MST156). | |||||
|
MYEF2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.093635 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C854, Q60690, Q6P930, Q6ZPT3, Q8QZZ1 | Gene names | Myef2, Kiaa1341, Mef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MEF-2). | |||||
|
PABP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.107243 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.046352 (rank : 85) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.071194 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PPIL4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.059224 (rank : 50) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
PPIL4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.059587 (rank : 47) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
HNRPM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.085565 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52272, Q15584, Q8WZ44, Q96H56, Q9BWL9, Q9Y492 | Gene names | HNRPM | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
HNRPM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.085930 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D0E1, Q6P1B2, Q99JQ0 | Gene names | Hnrpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
SRR35_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.110792 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |
|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
ST6B1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.016559 (rank : 89) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IMI4 | Gene names | SULT6B1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase 6B1 (EC 2.8.2.-). | |||||
|
GBRA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.006107 (rank : 90) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16445 | Gene names | GABRA6 | |||
|
Domain Architecture |
|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-6 precursor (GABA(A) receptor subunit alpha-6). | |||||
|
RBM14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.082861 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
RBM14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.082422 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.049774 (rank : 83) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.049774 (rank : 84) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.025797 (rank : 87) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
RBM19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.057743 (rank : 52) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
RNPC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.102293 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14498, Q14499 | Gene names | RNPC2, HCC1 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding region-containing protein 2 (Hepatocellular carcinoma protein 1) (Splicing factor HCC1). | |||||
|
RNPC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.102098 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VH51, Q99KV0 | Gene names | Rnpc2, Caper | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding region-containing protein 2 (Coactivator of activating protein 1 and estrogen receptors) (Coactivator of AP-1 and ERs) (Transcription coactivator CAPER). | |||||
|
ZC3H5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.018960 (rank : 88) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ACF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.051693 (rank : 72) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NQ94, Q5SZQ0, Q9NQ93, Q9NQX8, Q9NQX9, Q9NXC9, Q9NZD3 | Gene names | ACF, ASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
ACF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.050935 (rank : 76) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.052406 (rank : 65) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CHCH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.053278 (rank : 63) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NX63 | Gene names | CHCHD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3. | |||||
|
CLCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.064380 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09496 | Gene names | CLTA | |||
|
Domain Architecture |
|
|||||
| Description | Clathrin light chain A (Lca). | |||||
|
CLCB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.082572 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
CLCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.082840 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
CSTF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.065150 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
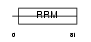
Domain Architecture |
|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
CSTF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.065699 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
CSTFT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.064570 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
CSTFT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.064051 (rank : 45) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
DND1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.052243 (rank : 67) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYX4 | Gene names | DND1, RBMS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dead end protein homolog 1 (RNA-binding motif, single-stranded- interacting protein 4). | |||||
|
DND1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.054330 (rank : 59) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6VY05, Q8CFK7 | Gene names | Dnd1, Rbms4, Ter | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dead end protein homolog 1 (RNA-binding motif, single-stranded- interacting protein 4). | |||||
|
FUSIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.097775 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
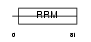
Domain Architecture |
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.097775 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
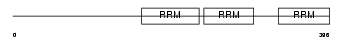
Domain Architecture |
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
HNRPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.051785 (rank : 69) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
HNRPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.052124 (rank : 68) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
HNRPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.053826 (rank : 61) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43390 | Gene names | HNRPR | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein R (hnRNP R). | |||||
|
MK67I_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.050898 (rank : 77) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BYG3, Q8TB66, Q96ED4 | Gene names | MKI67IP, NIFK, NOPP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MKI67 FHA domain-interacting nucleolar phosphoprotein (Nucleolar protein interacting with the FHA domain of pKI-67) (hNIFK) (Nucleolar phosphoprotein Nopp34). | |||||
|
NONO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.050470 (rank : 80) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |
|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050329 (rank : 81) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.059027 (rank : 51) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.053397 (rank : 62) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.050251 (rank : 82) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RB15B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.074385 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RB15B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.076030 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RBM28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.051218 (rank : 75) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NW13, Q96CV3 | Gene names | RBM28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
RBM7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051384 (rank : 73) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y580 | Gene names | RBM7 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
RBM7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.052850 (rank : 64) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CQT2, Q3UB91, Q7TQE3 | Gene names | Rbm7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
RBM8A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.070087 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y5S9, Q6FHD1, Q9GZX8, Q9NZI4 | Gene names | RBM8A, RBM8 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A) (RNA-binding protein Y14) (Binder of OVCA1- 1) (BOV-1). | |||||
|
RBM8A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.070087 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CWZ3, Q9D6U5 | Gene names | Rbm8a, Rbm8 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A). | |||||
|
RBMS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.051353 (rank : 74) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
RBMS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.052293 (rank : 66) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
RBMS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.050658 (rank : 79) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VC70 | Gene names | Rbms2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2. | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.062081 (rank : 46) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
REFP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.050886 (rank : 78) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJW6, Q8C869 | Gene names | Refbp2, Ref2 | |||
|
Domain Architecture |
|
|||||
| Description | RNA and export factor-binding protein 2. | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.059457 (rank : 49) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
STIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.059578 (rank : 48) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
STIM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.057295 (rank : 54) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
STMN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.051752 (rank : 70) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H169 | Gene names | STMN4 | |||
|
Domain Architecture |
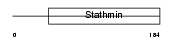
|
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
STMN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.051740 (rank : 71) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63042, O35414, O35415, O35416 | Gene names | Stmn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
TE2IP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.083690 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYB0, Q8WYZ3, Q9NWR2 | Gene names | TERF2IP, RAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1) (hRap1). | |||||
|
THOC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.055750 (rank : 57) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86V81, O43672 | Gene names | THOC4, ALY, BEF | |||
|
Domain Architecture |
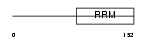
|
|||||
| Description | THO complex subunit 4 (Tho4) (Ally of AML-1 and LEF-1) (Transcriptional coactivator Aly/REF) (bZIP-enhancing factor BEF). | |||||
|
THOC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.056474 (rank : 56) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08583, Q8CBM4, Q9JJW7 | Gene names | Thoc4, Aly, Ref1, Refbp1 | |||
|
Domain Architecture |
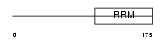
|
|||||
| Description | THO complex subunit 4 (Tho4) (RNA and export factor-binding protein 1) (REF1-I) (Ally of AML-1 and LEF-1) (Aly/REF). | |||||
|
TIA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.056689 (rank : 55) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P31483 | Gene names | TIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolysin TIA-1 isoform p40 (RNA-binding protein TIA-1) (p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15 (p15-TIA-1)]. | |||||
|
TIA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.057317 (rank : 53) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52912 | Gene names | Tia1, Tia | |||
|
Domain Architecture |
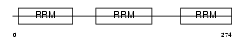
|
|||||
| Description | Nucleolysin TIA-1 (RNA-binding protein TIA-1). | |||||
|
TIAR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.055240 (rank : 58) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01085 | Gene names | TIAL1 | |||
|
Domain Architecture |
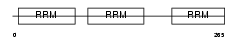
|
|||||
| Description | Nucleolysin TIAR (TIA-1-related protein). | |||||
|
TIAR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.054227 (rank : 60) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70318 | Gene names | Tial1 | |||
|
Domain Architecture |
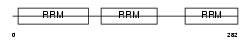
|
|||||
| Description | Nucleolysin TIAR (TIA-1-related protein). | |||||
|
U2AF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.072091 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26368 | Gene names | U2AF2, U2AF65 | |||
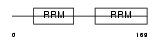
|
Domain Architecture |
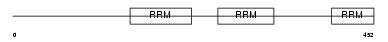
|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit) (hU2AF(65)). | |||||
|
U2AF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.072265 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26369 | Gene names | U2af2, U2af65 | |||
|
Domain Architecture |
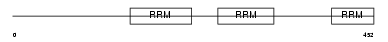
|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit). | |||||
|
U2AF1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.44581e-90 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01081 | Gene names | U2AF1, U2AF35 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
U2AF1_MOUSE
|
||||||
| NC score | 0.999707 (rank : 2) | θ value | 1.44581e-90 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D883, Q564E4, Q99LX2, Q9CZ98 | Gene names | U2af1 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
U2AFL_MOUSE
|
||||||
| NC score | 0.730026 (rank : 3) | θ value | 2.51237e-26 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.692101 (rank : 4) | θ value | 2.19584e-30 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.665524 (rank : 5) | θ value | 3.74554e-30 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.659222 (rank : 6) | θ value | 2.86786e-30 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
SPF45_HUMAN
|
||||||
| NC score | 0.212708 (rank : 7) | θ value | 0.00134147 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.212067 (rank : 8) | θ value | 0.00134147 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.126380 (rank : 9) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
PABP1_MOUSE
|
||||||
| NC score | 0.120431 (rank : 10) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP5_HUMAN
|
||||||
| NC score | 0.117978 (rank : 11) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96DU9, Q6P529, Q9UFE5 | Gene names | PABPC5, PABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 5 (Poly(A)-binding protein 5) (PABP 5). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.117579 (rank : 12) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
PABP1_HUMAN
|
||||||
| NC score | 0.117431 (rank : 13) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP3_HUMAN
|
||||||
| NC score | 0.117097 (rank : 14) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H361, Q8NHV0, Q9H086 | Gene names | PABPC3, PABP3, PABPL3 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 3 (Poly(A)-binding protein 3) (PABP 3) (Testis-specific poly(A)-binding protein). | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.112223 (rank : 15) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| NC score | 0.112001 (rank : 16) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
RBM4B_HUMAN
|
||||||
| NC score | 0.111938 (rank : 17) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| NC score | 0.111906 (rank : 18) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
SRR35_HUMAN
|
||||||
| NC score | 0.110792 (rank : 19) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |

|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
PABP4_HUMAN
|
||||||
| NC score | 0.107243 (rank : 20) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
RNPC2_HUMAN
|
||||||
| NC score | 0.102293 (rank : 21) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14498, Q14499 | Gene names | RNPC2, HCC1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 2 (Hepatocellular carcinoma protein 1) (Splicing factor HCC1). | |||||
|
RNPC2_MOUSE
|
||||||
| NC score | 0.102098 (rank : 22) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VH51, Q99KV0 | Gene names | Rnpc2, Caper | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 2 (Coactivator of activating protein 1 and estrogen receptors) (Coactivator of AP-1 and ERs) (Transcription coactivator CAPER). | |||||
|
FUSIP_HUMAN
|
||||||
| NC score | 0.097775 (rank : 23) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| NC score | 0.097775 (rank : 24) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
MYEF2_HUMAN
|
||||||
| NC score | 0.096352 (rank : 25) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2K5, Q6NUM5, Q7L388, Q7Z4B7, Q9H922, Q9NUQ1 | Gene names | MYEF2, KIAA1341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MST156). | |||||
|
MYEF2_MOUSE
|
||||||
| NC score | 0.093635 (rank : 26) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C854, Q60690, Q6P930, Q6ZPT3, Q8QZZ1 | Gene names | Myef2, Kiaa1341, Mef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MEF-2). | |||||
|
HNRPM_MOUSE
|
||||||
| NC score | 0.085930 (rank : 27) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D0E1, Q6P1B2, Q99JQ0 | Gene names | Hnrpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
HNRPM_HUMAN
|
||||||
| NC score | 0.085565 (rank : 28) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52272, Q15584, Q8WZ44, Q96H56, Q9BWL9, Q9Y492 | Gene names | HNRPM | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
TE2IP_HUMAN
|
||||||
| NC score | 0.083690 (rank : 29) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYB0, Q8WYZ3, Q9NWR2 | Gene names | TERF2IP, RAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1) (hRap1). | |||||
|
RBM14_HUMAN
|
||||||
| NC score | 0.082861 (rank : 30) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
CLCB_MOUSE
|
||||||
| NC score | 0.082840 (rank : 31) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
CLCB_HUMAN
|
||||||
| NC score | 0.082572 (rank : 32) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
RBM14_MOUSE
|
||||||
| NC score | 0.082422 (rank : 33) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.076030 (rank : 34) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RB15B_HUMAN
|
||||||
| NC score | 0.074385 (rank : 35) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
U2AF2_MOUSE
|
||||||
| NC score | 0.072265 (rank : 36) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26369 | Gene names | U2af2, U2af65 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit). | |||||
|
U2AF2_HUMAN
|
||||||
| NC score | 0.072091 (rank : 37) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26368 | Gene names | U2AF2, U2AF65 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit) (hU2AF(65)). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.071194 (rank : 38) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RBM8A_HUMAN
|
||||||
| NC score | 0.070087 (rank : 39) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y5S9, Q6FHD1, Q9GZX8, Q9NZI4 | Gene names | RBM8A, RBM8 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A) (RNA-binding protein Y14) (Binder of OVCA1- 1) (BOV-1). | |||||
|
RBM8A_MOUSE
|
||||||
| NC score | 0.070087 (rank : 40) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CWZ3, Q9D6U5 | Gene names | Rbm8a, Rbm8 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A). | |||||
|
CSTF2_MOUSE
|
||||||
| NC score | 0.065699 (rank : 41) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
CSTF2_HUMAN
|
||||||
| NC score | 0.065150 (rank : 42) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
CSTFT_HUMAN
|
||||||
| NC score | 0.064570 (rank : 43) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
CLCA_HUMAN
|
||||||
| NC score | 0.064380 (rank : 44) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09496 | Gene names | CLTA | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain A (Lca). | |||||
|
CSTFT_MOUSE
|
||||||
| NC score | 0.064051 (rank : 45) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.062081 (rank : 46) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
PPIL4_MOUSE
|
||||||
| NC score | 0.059587 (rank : 47) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.059578 (rank : 48) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.059457 (rank : 49) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
PPIL4_HUMAN
|
||||||
| NC score | 0.059224 (rank : 50) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.059027 (rank : 51) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RBM19_HUMAN
|
||||||
| NC score | 0.057743 (rank : 52) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
TIA1_MOUSE
|
||||||
| NC score | 0.057317 (rank : 53) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52912 | Gene names | Tia1, Tia | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolysin TIA-1 (RNA-binding protein TIA-1). | |||||
|
STIM1_MOUSE
|
||||||
| NC score | 0.057295 (rank : 54) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TIA1_HUMAN
|
||||||
| NC score | 0.056689 (rank : 55) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P31483 | Gene names | TIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolysin TIA-1 isoform p40 (RNA-binding protein TIA-1) (p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15 (p15-TIA-1)]. | |||||
|
THOC4_MOUSE
|
||||||
| NC score | 0.056474 (rank : 56) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08583, Q8CBM4, Q9JJW7 | Gene names | Thoc4, Aly, Ref1, Refbp1 | |||
|
Domain Architecture |

|
|||||
| Description | THO complex subunit 4 (Tho4) (RNA and export factor-binding protein 1) (REF1-I) (Ally of AML-1 and LEF-1) (Aly/REF). | |||||
|
THOC4_HUMAN
|
||||||
| NC score | 0.055750 (rank : 57) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86V81, O43672 | Gene names | THOC4, ALY, BEF | |||
|
Domain Architecture |

|
|||||
| Description | THO complex subunit 4 (Tho4) (Ally of AML-1 and LEF-1) (Transcriptional coactivator Aly/REF) (bZIP-enhancing factor BEF). | |||||
|
TIAR_HUMAN
|
||||||
| NC score | 0.055240 (rank : 58) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01085 | Gene names | TIAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolysin TIAR (TIA-1-related protein). | |||||
|
DND1_MOUSE
|
||||||
| NC score | 0.054330 (rank : 59) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6VY05, Q8CFK7 | Gene names | Dnd1, Rbms4, Ter | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dead end protein homolog 1 (RNA-binding motif, single-stranded- interacting protein 4). | |||||
|
TIAR_MOUSE
|
||||||
| NC score | 0.054227 (rank : 60) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70318 | Gene names | Tial1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolysin TIAR (TIA-1-related protein). | |||||
|
HNRPR_HUMAN
|
||||||
| NC score | 0.053826 (rank : 61) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43390 | Gene names | HNRPR | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein R (hnRNP R). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.053397 (rank : 62) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
CHCH3_HUMAN
|
||||||
| NC score | 0.053278 (rank : 63) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NX63 | Gene names | CHCHD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3. | |||||
|
RBM7_MOUSE
|
||||||
| NC score | 0.052850 (rank : 64) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CQT2, Q3UB91, Q7TQE3 | Gene names | Rbm7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.052406 (rank : 65) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
RBMS1_MOUSE
|
||||||
| NC score | 0.052293 (rank : 66) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
DND1_HUMAN
|
||||||
| NC score | 0.052243 (rank : 67) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYX4 | Gene names | DND1, RBMS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dead end protein homolog 1 (RNA-binding motif, single-stranded- interacting protein 4). | |||||
|
HNRPQ_MOUSE
|
||||||
| NC score | 0.052124 (rank : 68) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
HNRPQ_HUMAN
|
||||||
| NC score | 0.051785 (rank : 69) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
STMN4_HUMAN
|
||||||
| NC score | 0.051752 (rank : 70) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H169 | Gene names | STMN4 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
STMN4_MOUSE
|
||||||
| NC score | 0.051740 (rank : 71) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63042, O35414, O35415, O35416 | Gene names | Stmn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
ACF_HUMAN
|
||||||
| NC score | 0.051693 (rank : 72) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NQ94, Q5SZQ0, Q9NQ93, Q9NQX8, Q9NQX9, Q9NXC9, Q9NZD3 | Gene names | ACF, ASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
RBM7_HUMAN
|
||||||
| NC score | 0.051384 (rank : 73) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y580 | Gene names | RBM7 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
RBMS1_HUMAN
|
||||||
| NC score | 0.051353 (rank : 74) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
RBM28_HUMAN
|
||||||
| NC score | 0.051218 (rank : 75) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NW13, Q96CV3 | Gene names | RBM28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
ACF_MOUSE
|
||||||
| NC score | 0.050935 (rank : 76) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
MK67I_HUMAN
|
||||||
| NC score | 0.050898 (rank : 77) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BYG3, Q8TB66, Q96ED4 | Gene names | MKI67IP, NIFK, NOPP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MKI67 FHA domain-interacting nucleolar phosphoprotein (Nucleolar protein interacting with the FHA domain of pKI-67) (hNIFK) (Nucleolar phosphoprotein Nopp34). | |||||
|
REFP2_MOUSE
|
||||||
| NC score | 0.050886 (rank : 78) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJW6, Q8C869 | Gene names | Refbp2, Ref2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA and export factor-binding protein 2. | |||||
|
RBMS2_MOUSE
|
||||||
| NC score | 0.050658 (rank : 79) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VC70 | Gene names | Rbms2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2. | |||||
|
NONO_HUMAN
|
||||||
| NC score | 0.050470 (rank : 80) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |

|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| NC score | 0.050329 (rank : 81) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.050251 (rank : 82) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.049774 (rank : 83) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.049774 (rank : 84) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.046352 (rank : 85) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.039165 (rank : 86) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.025797 (rank : 87) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
ZC3H5_MOUSE
|
||||||
| NC score | 0.018960 (rank : 88) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ST6B1_HUMAN
|
||||||
| NC score | 0.016559 (rank : 89) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IMI4 | Gene names | SULT6B1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase 6B1 (EC 2.8.2.-). | |||||
|
GBRA6_HUMAN
|
||||||
| NC score | 0.006107 (rank : 90) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16445 | Gene names | GABRA6 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-6 precursor (GABA(A) receptor subunit alpha-6). | |||||