Please be patient as the page loads
|
TF3C1_HUMAN
|
||||||
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TF3C1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
TF3C1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.973778 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K284, Q8CAL9 | Gene names | Gtf3c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
WDR22_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 3) | NC score | 0.057933 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |
|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
RNF25_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 4) | NC score | 0.063826 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96BH1, Q9H874 | Gene names | RNF25 | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-). | |||||
|
S6A12_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.021608 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31651 | Gene names | Slc6a12, Gabt2, Gat-2, Gat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent betaine transporter (Na+/Cl- betaine/GABA transporter) (Sodium- and chloride-dependent GABA transporter 2) (GAT2). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 6) | NC score | 0.052349 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.029517 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.031741 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.031243 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GFAP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.018178 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
PPIL2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.046774 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13356, Q13357, Q8TAH2, Q9BWR8 | Gene names | PPIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.026458 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.018064 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
FA98B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.054388 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q52LJ0, Q8N935 | Gene names | FAM98B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98B. | |||||
|
IF16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.036068 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
NGEF_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.027677 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.029539 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PPIL2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.044892 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D787, Q542A2, Q9CZL1 | Gene names | Ppil2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.028445 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.048169 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
WEE1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.011692 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q66JT0, Q4U4S4, Q7TPV9 | Gene names | Wee1b, Wee2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase) (mWee1B). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.015451 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DCX_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.022948 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.045521 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.048985 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LIPB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.020682 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.034508 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
EDRF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.035811 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GQV7, Q80ZY3, Q8R3P5 | Gene names | Edrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
FA8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.015562 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.012579 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.006107 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.021237 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.039570 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.034609 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.010562 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
VATD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.033703 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P57746 | Gene names | Atp6v1d, Atp6m, Vatd | |||
|
Domain Architecture |
|
|||||
| Description | Vacuolar ATP synthase subunit D (EC 3.6.3.14) (V-ATPase D subunit) (Vacuolar proton pump D subunit) (V-ATPase 28 kDa accessory protein). | |||||
|
ZSCA5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.003245 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
PDZK4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.015857 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76G19, Q8NB75, Q9BUH9, Q9P284 | Gene names | PDZK4, KIAA1444, PDZRN4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.011845 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
RNF31_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.019431 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.014318 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.007473 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CO6A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.012409 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |
|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.021894 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ZBT17_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.001601 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
BIM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.022968 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54918, O54919, O54920 | Gene names | Bcl2l11, Bim | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
DNAS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.013826 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49183, O70532 | Gene names | Dnase1, Dnl1 | |||
|
Domain Architecture |
|
|||||
| Description | Deoxyribonuclease-1 precursor (EC 3.1.21.1) (Deoxyribonuclease I) (DNase I). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.009627 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NIBL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.016910 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96TA1, Q9BUS2, Q9NT35 | Gene names | C9orf88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein (Meg-3). | |||||
|
PI3R5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.018204 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SW28, Q3TU01, Q3UDZ2, Q8C215, Q8CGQ7 | Gene names | Pik3r5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 5 (PI3-kinase regulatory subunit 5) (PI3-kinase p101 subunit) (PtdIns-3-kinase p101) (p101- PI3K) (Phosphatidylinositol-4,5-bisphosphate 3-kinase regulatory subunit) (PtdIns-3-kinase regulatory subunit). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.035767 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.025519 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.011119 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.023181 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.018781 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.022717 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.022062 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
ATS12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.006508 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q811B3, Q8BK92, Q8BKY1 | Gene names | Adamts12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.011569 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
EDRF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.028504 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3B7T1, Q4G190, Q8IZ74, Q9Y3W4 | Gene names | EDRF1, C10orf137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.025183 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.023209 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MPRI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.012771 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
MTF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.001967 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
NFIP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.019314 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NV92, Q7Z2H3, Q7Z428, Q8TAR3, Q9ULQ5 | Gene names | NDFIP2, KIAA1165, N4WBP5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD4 family-interacting protein 2 (NEDD4 WW domain-binding protein 5A) (Putative MAPK-activating protein PM04/PM05/PM06/PM07) (Putative NF-kappa-B-activating protein 413). | |||||
|
PARN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.016530 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95453 | Gene names | PARN, DAN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A)-specific ribonuclease PARN (EC 3.1.13.4) (Polyadenylate- specific ribonuclease) (Deadenylating nuclease) (Deadenylation nuclease). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.016628 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.012526 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.005489 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.001496 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.005579 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATS12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.005959 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.009070 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CCD76_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.013855 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYH3 | Gene names | Ccdc76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 76. | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.016939 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
GFAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.013473 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
GTF2I_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.011364 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
KBTBA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.006646 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60662 | Gene names | KBTBD10, KRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 10 (Kelch-related protein 1) (Kel-like protein 23) (Sarcosin). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.012793 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.010624 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
PO3F1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.004249 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21952 | Gene names | Pou3f1, Oct6, Otf-6, Otf6, Scip | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 1 (Octamer-binding transcription factor 6) (Oct-6) (POU-domain transcription factor SCIP). | |||||
|
RS9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.021723 (rank : 36) | |||
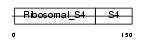
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46781, Q4QRK7, Q9BVZ0 | Gene names | RPS9 | |||
|
Domain Architecture |
|
|||||
| Description | 40S ribosomal protein S9. | |||||
|
RS9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.021723 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZWN5, Q3UBF1, Q8K2D1 | Gene names | Rps9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 40S ribosomal protein S9. | |||||
|
S6A11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.014845 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31650 | Gene names | Slc6a11, Gabt4, Gat-4, Gat4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent GABA transporter 4 (GAT4). | |||||
|
SC6A5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.012121 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.020833 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TBCD8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.009547 (rank : 75) | |||
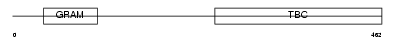
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1A9 | Gene names | Tbc1d8, Hblp1, Vrp | |||
|
Domain Architecture |
|
|||||
| Description | TBC1 domain family member 8 (Vascular Rab-GAP/TBC-containing protein) (BUB2-like protein 1). | |||||
|
ZN365_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.015187 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
TF3C1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
TF3C1_MOUSE
|
||||||
| NC score | 0.973778 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K284, Q8CAL9 | Gene names | Gtf3c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
RNF25_HUMAN
|
||||||
| NC score | 0.063826 (rank : 3) | θ value | 0.0736092 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96BH1, Q9H874 | Gene names | RNF25 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-). | |||||
|
WDR22_HUMAN
|
||||||
| NC score | 0.057933 (rank : 4) | θ value | 0.00175202 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
FA98B_HUMAN
|
||||||
| NC score | 0.054388 (rank : 5) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q52LJ0, Q8N935 | Gene names | FAM98B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98B. | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.052349 (rank : 6) | θ value | 0.125558 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.048985 (rank : 7) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.048169 (rank : 8) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
PPIL2_HUMAN
|
||||||
| NC score | 0.046774 (rank : 9) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13356, Q13357, Q8TAH2, Q9BWR8 | Gene names | PPIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.045521 (rank : 10) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
PPIL2_MOUSE
|
||||||
| NC score | 0.044892 (rank : 11) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D787, Q542A2, Q9CZL1 | Gene names | Ppil2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.039570 (rank : 12) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.036068 (rank : 13) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
EDRF1_MOUSE
|
||||||
| NC score | 0.035811 (rank : 14) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GQV7, Q80ZY3, Q8R3P5 | Gene names | Edrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.035767 (rank : 15) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.034609 (rank : 16) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.034508 (rank : 17) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
VATD_MOUSE
|
||||||
| NC score | 0.033703 (rank : 18) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P57746 | Gene names | Atp6v1d, Atp6m, Vatd | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar ATP synthase subunit D (EC 3.6.3.14) (V-ATPase D subunit) (Vacuolar proton pump D subunit) (V-ATPase 28 kDa accessory protein). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.031741 (rank : 19) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.031243 (rank : 20) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.029539 (rank : 21) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.029517 (rank : 22) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
EDRF1_HUMAN
|
||||||
| NC score | 0.028504 (rank : 23) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3B7T1, Q4G190, Q8IZ74, Q9Y3W4 | Gene names | EDRF1, C10orf137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.028445 (rank : 24) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
NGEF_MOUSE
|
||||||
| NC score | 0.027677 (rank : 25) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.026458 (rank : 26) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.025519 (rank : 27) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.025183 (rank : 28) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.023209 (rank : 29) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.023181 (rank : 30) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BIM_MOUSE
|
||||||
| NC score | 0.022968 (rank : 31) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54918, O54919, O54920 | Gene names | Bcl2l11, Bim | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.022948 (rank : 32) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.022717 (rank : 33) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.022062 (rank : 34) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.021894 (rank : 35) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RS9_HUMAN
|
||||||
| NC score | 0.021723 (rank : 36) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46781, Q4QRK7, Q9BVZ0 | Gene names | RPS9 | |||
|
Domain Architecture |

|
|||||
| Description | 40S ribosomal protein S9. | |||||
|
RS9_MOUSE
|
||||||
| NC score | 0.021723 (rank : 37) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZWN5, Q3UBF1, Q8K2D1 | Gene names | Rps9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 40S ribosomal protein S9. | |||||
|
S6A12_MOUSE
|
||||||
| NC score | 0.021608 (rank : 38) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31651 | Gene names | Slc6a12, Gabt2, Gat-2, Gat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent betaine transporter (Na+/Cl- betaine/GABA transporter) (Sodium- and chloride-dependent GABA transporter 2) (GAT2). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.021237 (rank : 39) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.020833 (rank : 40) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
LIPB2_MOUSE
|
||||||
| NC score | 0.020682 (rank : 41) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
RNF31_MOUSE
|
||||||
| NC score | 0.019431 (rank : 42) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
NFIP2_HUMAN
|
||||||
| NC score | 0.019314 (rank : 43) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NV92, Q7Z2H3, Q7Z428, Q8TAR3, Q9ULQ5 | Gene names | NDFIP2, KIAA1165, N4WBP5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD4 family-interacting protein 2 (NEDD4 WW domain-binding protein 5A) (Putative MAPK-activating protein PM04/PM05/PM06/PM07) (Putative NF-kappa-B-activating protein 413). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.018781 (rank : 44) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
PI3R5_MOUSE
|
||||||
| NC score | 0.018204 (rank : 45) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SW28, Q3TU01, Q3UDZ2, Q8C215, Q8CGQ7 | Gene names | Pik3r5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 5 (PI3-kinase regulatory subunit 5) (PI3-kinase p101 subunit) (PtdIns-3-kinase p101) (p101- PI3K) (Phosphatidylinositol-4,5-bisphosphate 3-kinase regulatory subunit) (PtdIns-3-kinase regulatory subunit). | |||||
|
GFAP_MOUSE
|
||||||
| NC score | 0.018178 (rank : 46) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.018064 (rank : 47) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.016939 (rank : 48) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
NIBL_HUMAN
|
||||||
| NC score | 0.016910 (rank : 49) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96TA1, Q9BUS2, Q9NT35 | Gene names | C9orf88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein (Meg-3). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.016628 (rank : 50) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
PARN_HUMAN
|
||||||
| NC score | 0.016530 (rank : 51) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95453 | Gene names | PARN, DAN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A)-specific ribonuclease PARN (EC 3.1.13.4) (Polyadenylate- specific ribonuclease) (Deadenylating nuclease) (Deadenylation nuclease). | |||||
|
PDZK4_HUMAN
|
||||||
| NC score | 0.015857 (rank : 52) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76G19, Q8NB75, Q9BUH9, Q9P284 | Gene names | PDZK4, KIAA1444, PDZRN4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
FA8_MOUSE
|
||||||
| NC score | 0.015562 (rank : 53) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.015451 (rank : 54) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
ZN365_HUMAN
|
||||||
| NC score | 0.015187 (rank : 55) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
S6A11_MOUSE
|
||||||
| NC score | 0.014845 (rank : 56) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31650 | Gene names | Slc6a11, Gabt4, Gat-4, Gat4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent GABA transporter 4 (GAT4). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.014318 (rank : 57) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
CCD76_MOUSE
|
||||||
| NC score | 0.013855 (rank : 58) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYH3 | Gene names | Ccdc76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 76. | |||||
|
DNAS1_MOUSE
|
||||||
| NC score | 0.013826 (rank : 59) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49183, O70532 | Gene names | Dnase1, Dnl1 | |||
|
Domain Architecture |
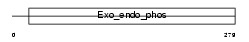
|
|||||
| Description | Deoxyribonuclease-1 precursor (EC 3.1.21.1) (Deoxyribonuclease I) (DNase I). | |||||
|
GFAP_HUMAN
|
||||||
| NC score | 0.013473 (rank : 60) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.012793 (rank : 61) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MPRI_MOUSE
|
||||||
| NC score | 0.012771 (rank : 62) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.012579 (rank : 63) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.012526 (rank : 64) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
CO6A1_HUMAN
|
||||||
| NC score | 0.012409 (rank : 65) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
SC6A5_MOUSE
|
||||||
| NC score | 0.012121 (rank : 66) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.011845 (rank : 67) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
WEE1B_MOUSE
|
||||||
| NC score | 0.011692 (rank : 68) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q66JT0, Q4U4S4, Q7TPV9 | Gene names | Wee1b, Wee2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase) (mWee1B). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.011569 (rank : 69) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
GTF2I_HUMAN
|
||||||
| NC score | 0.011364 (rank : 70) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.011119 (rank : 71) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.010624 (rank : 72) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.010562 (rank : 73) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.009627 (rank : 74) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
TBCD8_MOUSE
|
||||||
| NC score | 0.009547 (rank : 75) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1A9 | Gene names | Tbc1d8, Hblp1, Vrp | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 8 (Vascular Rab-GAP/TBC-containing protein) (BUB2-like protein 1). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.009070 (rank : 76) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.007473 (rank : 77) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
KBTBA_HUMAN
|
||||||
| NC score | 0.006646 (rank : 78) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60662 | Gene names | KBTBD10, KRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 10 (Kelch-related protein 1) (Kel-like protein 23) (Sarcosin). | |||||
|
ATS12_MOUSE
|
||||||
| NC score | 0.006508 (rank : 79) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q811B3, Q8BK92, Q8BKY1 | Gene names | Adamts12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.006107 (rank : 80) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
ATS12_HUMAN
|
||||||
| NC score | 0.005959 (rank : 81) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.005579 (rank : 82) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.005489 (rank : 83) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
PO3F1_MOUSE
|
||||||
| NC score | 0.004249 (rank : 84) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21952 | Gene names | Pou3f1, Oct6, Otf-6, Otf6, Scip | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 1 (Octamer-binding transcription factor 6) (Oct-6) (POU-domain transcription factor SCIP). | |||||
|
ZSCA5_HUMAN
|
||||||
| NC score | 0.003245 (rank : 85) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
MTF1_HUMAN
|
||||||
| NC score | 0.001967 (rank : 86) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
ZBT17_MOUSE
|
||||||
| NC score | 0.001601 (rank : 87) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | 0.001496 (rank : 88) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||