Please be patient as the page loads
|
FA8_MOUSE
|
||||||
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FA8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989977 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
FA8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 1.12453e-151 (rank : 3) | NC score | 0.932030 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
HEPH_MOUSE
|
||||||
| θ value | 7.33985e-127 (rank : 4) | NC score | 0.699447 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
HEPH_HUMAN
|
||||||
| θ value | 7.35691e-119 (rank : 5) | NC score | 0.696814 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
CERU_HUMAN
|
||||||
| θ value | 1.53401e-116 (rank : 6) | NC score | 0.697488 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
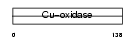
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
CERU_MOUSE
|
||||||
| θ value | 4.32305e-111 (rank : 7) | NC score | 0.695125 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
EDIL3_HUMAN
|
||||||
| θ value | 3.70236e-70 (rank : 8) | NC score | 0.551348 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
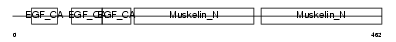
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| θ value | 8.24801e-70 (rank : 9) | NC score | 0.542624 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
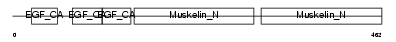
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
MFGM_MOUSE
|
||||||
| θ value | 3.24342e-66 (rank : 10) | NC score | 0.666579 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |
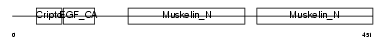
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
MFGM_HUMAN
|
||||||
| θ value | 5.92465e-60 (rank : 11) | NC score | 0.766711 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |
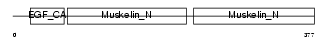
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
NRP2_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 12) | NC score | 0.539585 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP1_MOUSE
|
||||||
| θ value | 4.71619e-41 (rank : 13) | NC score | 0.532854 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
NRP2_HUMAN
|
||||||
| θ value | 6.15952e-41 (rank : 14) | NC score | 0.537107 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP1_HUMAN
|
||||||
| θ value | 1.08726e-37 (rank : 15) | NC score | 0.529884 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 16) | NC score | 0.576101 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
DCBD2_MOUSE
|
||||||
| θ value | 1.62847e-25 (rank : 17) | NC score | 0.587725 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |
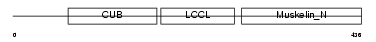
|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD1_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 18) | NC score | 0.586888 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
CPXM2_MOUSE
|
||||||
| θ value | 1.29031e-22 (rank : 19) | NC score | 0.482075 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_HUMAN
|
||||||
| θ value | 4.9032e-22 (rank : 20) | NC score | 0.479324 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
XLRS1_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 21) | NC score | 0.729120 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |
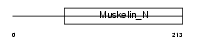
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
XLRS1_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 22) | NC score | 0.728251 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |
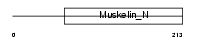
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
CPXM1_MOUSE
|
||||||
| θ value | 1.38178e-16 (rank : 23) | NC score | 0.435240 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CNTP2_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 24) | NC score | 0.461825 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CPXM1_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 25) | NC score | 0.442625 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CNTP4_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 26) | NC score | 0.444602 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP2_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 27) | NC score | 0.449439 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP4_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 28) | NC score | 0.441851 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP3_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 29) | NC score | 0.452176 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 30) | NC score | 0.420260 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
DDR2_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 31) | NC score | 0.161934 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62371 | Gene names | Ddr2, Ntrkr3, Tkt, Tyro10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
DDR2_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 32) | NC score | 0.160189 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16832 | Gene names | DDR2, NTRKR3, TKT, TYRO10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
CNTP1_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 33) | NC score | 0.418377 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
DDR1_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 34) | NC score | 0.148327 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08345, Q14196, Q16562, Q5ST12, Q9UD37 | Gene names | DDR1, CAK, EDDR1, RTK6, TRKE | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (TRK E) (Protein-tyrosine kinase RTK 6) (HGK2) (CD167a antigen). | |||||
|
DDR1_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 35) | NC score | 0.141448 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03146 | Gene names | Ddr1, Cak, Eddr1, Mpk6 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (Protein-tyrosine kinase MPK-6) (CD167a antigen). | |||||
|
SMC4_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 36) | NC score | 0.043829 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 37) | NC score | 0.036384 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 38) | NC score | 0.013908 (rank : 122) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
RIF1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 39) | NC score | 0.028172 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.046316 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
UBP2L_MOUSE
|
||||||
| θ value | 0.125558 (rank : 41) | NC score | 0.054557 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
NRIP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.033699 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
TERF2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.037582 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.040531 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.018139 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
UBP2L_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.050703 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |
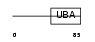
|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.039479 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
XTP3B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.036085 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DZ1, O95901, Q6UWN7, Q9NUY7, Q9UQL4 | Gene names | C2orf30, XTP3TPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XTP3-transactivated gene B protein precursor. | |||||
|
AKAP5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.035465 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24588 | Gene names | AKAP5, AKAP79 | |||
|
Domain Architecture |
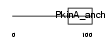
|
|||||
| Description | A-kinase anchor protein 5 (A-kinase anchor protein 79 kDa) (AKAP 79) (cAMP-dependent protein kinase regulatory subunit II high affinity- binding protein) (H21). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.016979 (rank : 116) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.016989 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
XTP3B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.035043 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEH8, Q3UZF3, Q8BVN6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XTP3-transactivated gene B protein homolog precursor. | |||||
|
TBCD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.014642 (rank : 121) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.008881 (rank : 132) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.015808 (rank : 118) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.020012 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
CA026_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.018101 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.016538 (rank : 117) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
TARA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.012817 (rank : 123) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.010617 (rank : 129) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.011259 (rank : 128) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.011304 (rank : 127) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.015182 (rank : 120) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
MNT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.018848 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.011965 (rank : 125) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
TERF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.025412 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54274, Q15553, Q93029 | Gene names | TERF1, PIN2, TRF, TRF1 | |||
|
Domain Architecture |
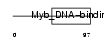
|
|||||
| Description | Telomeric repeat-binding factor 1 (TTAGGG repeat-binding factor 1) (NIMA-interacting protein 2) (Telomeric protein Pin2/TRF1). | |||||
|
TF3C1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.015562 (rank : 119) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
K0082_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.020479 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K1C13_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.003802 (rank : 147) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P13646, Q53G54, Q6AZK5, Q8N240 | Gene names | KRT13 | |||
|
Domain Architecture |
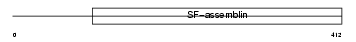
|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13). | |||||
|
TERF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.039225 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |
|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TOM70_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.018813 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.020057 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.006696 (rank : 139) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.012592 (rank : 124) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.011904 (rank : 126) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PLK4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.002894 (rank : 151) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64702, Q6PEP6, Q78EG6, Q8R0I5, Q9CVR6, Q9CVU6 | Gene names | Plk4, Sak, Stk18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.005309 (rank : 144) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
ZHX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.006532 (rank : 140) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
CP135_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.008905 (rank : 131) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.006521 (rank : 141) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
ICOSL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.009109 (rank : 130) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75144, Q9HD18, Q9NRQ1 | Gene names | ICOSLG, B7H2, B7RP1, ICOSL, KIAA0653 | |||
|
Domain Architecture |
|
|||||
| Description | ICOS ligand precursor (B7 homolog 2) (B7-H2) (B7-like protein Gl50) (B7-related protein 1) (B7RP-1) (CD275 antigen). | |||||
|
NRIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.023258 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
REST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.006775 (rank : 138) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
K1C13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.003298 (rank : 150) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08730, Q3V176, Q6PAI1 | Gene names | Krt13, Krt1-13 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13) (47 kDa cytokeratin). | |||||
|
KIF3B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.003522 (rank : 149) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
RN168_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.004314 (rank : 145) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
TM117_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.033195 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BH18, Q8C6X8 | Gene names | Tmem117 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 117. | |||||
|
ZBT7B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | -0.004187 (rank : 154) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15156, Q68DR2, Q96EP2 | Gene names | ZBTB7B, ZBTB15, ZFP67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 7B (Zinc finger protein 67 homolog) (Zfp-67) (Zinc finger protein Th-POK) (T-helper-inducing POZ/Krueppel-like factor) (Krueppel-related zinc finger protein cKrox) (hcKrox) (Zinc finger and BTB domain-containing protein 15). | |||||
|
ZN578_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | -0.004489 (rank : 155) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96N58 | Gene names | ZNF578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 578. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.005584 (rank : 142) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.005335 (rank : 143) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GFAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.003951 (rank : 146) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.007223 (rank : 136) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
PLD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.008229 (rank : 133) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13393 | Gene names | PLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choline phosphatase 1) (Phosphatidylcholine-hydrolyzing phospholipase D1) (hPLD1). | |||||
|
PLD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.008166 (rank : 134) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z280, O35911 | Gene names | Pld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choline phosphatase 1) (Phosphatidylcholine-hydrolyzing phospholipase D1) (mPLD1). | |||||
|
PODXL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.007367 (rank : 135) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0M4, Q9ESZ1 | Gene names | Podxl, Pclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
SPB12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.000498 (rank : 153) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7P9, Q6UKZ3 | Gene names | Serpinb12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B12. | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.002514 (rank : 152) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
TRPM6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.003533 (rank : 148) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TTC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.007019 (rank : 137) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.064007 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
BMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.075857 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.075747 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
CBPD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.087576 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75976, O15377, Q86SH9, Q86XE6 | Gene names | CPD | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.087566 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89001 | Gene names | Cpd | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.101604 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16870, Q9UIU9 | Gene names | CPE | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.101487 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.091017 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14384, Q9H2K9 | Gene names | CPM | |||
|
Domain Architecture |
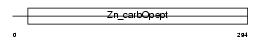
|
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.091368 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V42, Q497S5, Q9CYH8 | Gene names | Cpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.099891 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15169 | Gene names | CPN1, ACBP | |||
|
Domain Architecture |
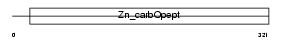
|
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit) (Lysine carboxypeptidase) (Arginine carboxypeptidase) (Kininase-1) (Serum carboxypeptidase N) (SCPN) (Anaphylatoxin inactivator) (Plasma carboxypeptidase B). | |||||
|
CBPN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.100235 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
CBPZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.089221 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.090574 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CRUM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.051716 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.052456 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.052666 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CSMD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.052321 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056301 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
CSMD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.053220 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.087115 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.087610 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.087792 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.082402 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
DCBD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.157882 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.065125 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
GLPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.099204 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |
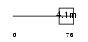
|
|||||
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
GP126_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.054820 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
LRP12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.056176 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |
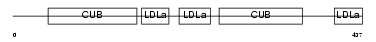
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
LRP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.058237 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |
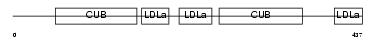
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
MAMC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.068641 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z304, Q5VW47, Q8WX43, Q96BM4 | Gene names | MAMDC2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
MAMC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.063999 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG85, Q8BM24 | Gene names | Mamdc2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
MFRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.079951 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.081092 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NETO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.095723 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.098321 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.101660 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.099674 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NRX1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.090912 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.089870 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CS84, O88722, Q80Y87, Q8CHE6 | Gene names | Nrxn1, Kiaa0578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.077069 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.087718 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4C0, O95378, Q9NS47, Q9P1V3, Q9P1V6, Q9UIE2, Q9UIE3, Q9ULA5, Q9Y486 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |
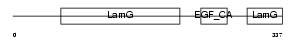
|
|||||
| Description | Neurexin-3-alpha precursor (Neurexin III-alpha). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.112364 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.114208 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.111528 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.114280 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PDGFD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.088712 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
PDGFD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.074897 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.060786 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
SHBG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.053035 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04278, P14689, Q16616 | Gene names | SHBG | |||
|
Domain Architecture |
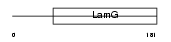
|
|||||
| Description | Sex hormone-binding globulin precursor (SHBG) (Sex steroid-binding protein) (SBP) (Testis-specific androgen-binding protein) (ABP) (Testosterone-estrogen-binding globulin) (Testosterone-estradiol- binding globulin) (TeBG). | |||||
|
TLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.074952 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.074081 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.074937 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.075473 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.096279 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.096797 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
FA8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.989977 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.932030 (rank : 3) | θ value | 1.12453e-151 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
MFGM_HUMAN
|
||||||
| NC score | 0.766711 (rank : 4) | θ value | 5.92465e-60 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
XLRS1_MOUSE
|
||||||
| NC score | 0.729120 (rank : 5) | θ value | 2.06002e-20 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
XLRS1_HUMAN
|
||||||
| NC score | 0.728251 (rank : 6) | θ value | 7.82807e-20 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
HEPH_MOUSE
|
||||||
| NC score | 0.699447 (rank : 7) | θ value | 7.33985e-127 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
CERU_HUMAN
|
||||||
| NC score | 0.697488 (rank : 8) | θ value | 1.53401e-116 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
HEPH_HUMAN
|
||||||
| NC score | 0.696814 (rank : 9) | θ value | 7.35691e-119 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
CERU_MOUSE
|
||||||
| NC score | 0.695125 (rank : 10) | θ value | 4.32305e-111 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
MFGM_MOUSE
|
||||||
| NC score | 0.666579 (rank : 11) | θ value | 3.24342e-66 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
DCBD2_MOUSE
|
||||||
| NC score | 0.587725 (rank : 12) | θ value | 1.62847e-25 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD1_HUMAN
|
||||||
| NC score | 0.586888 (rank : 13) | θ value | 4.43474e-23 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.576101 (rank : 14) | θ value | 3.87602e-27 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
EDIL3_HUMAN
|
||||||
| NC score | 0.551348 (rank : 15) | θ value | 3.70236e-70 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
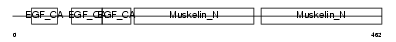
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| NC score | 0.542624 (rank : 16) | θ value | 8.24801e-70 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
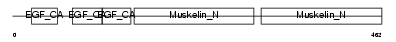
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
NRP2_MOUSE
|
||||||
| NC score | 0.539585 (rank : 17) | θ value | 2.11701e-41 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_HUMAN
|
||||||
| NC score | 0.537107 (rank : 18) | θ value | 6.15952e-41 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP1_MOUSE
|
||||||
| NC score | 0.532854 (rank : 19) | θ value | 4.71619e-41 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
NRP1_HUMAN
|
||||||
| NC score | 0.529884 (rank : 20) | θ value | 1.08726e-37 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
CPXM2_MOUSE
|
||||||
| NC score | 0.482075 (rank : 21) | θ value | 1.29031e-22 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_HUMAN
|
||||||
| NC score | 0.479324 (rank : 22) | θ value | 4.9032e-22 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CNTP2_HUMAN
|
||||||
| NC score | 0.461825 (rank : 23) | θ value | 5.25075e-16 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP3_HUMAN
|
||||||
| NC score | 0.452176 (rank : 24) | θ value | 6.41864e-14 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
CNTP2_MOUSE
|
||||||
| NC score | 0.449439 (rank : 25) | θ value | 2.20605e-14 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP4_MOUSE
|
||||||
| NC score | 0.444602 (rank : 26) | θ value | 1.68911e-14 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CPXM1_HUMAN
|
||||||
| NC score | 0.442625 (rank : 27) | θ value | 1.52774e-15 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CNTP4_HUMAN
|
||||||
| NC score | 0.441851 (rank : 28) | θ value | 2.88119e-14 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CPXM1_MOUSE
|
||||||
| NC score | 0.435240 (rank : 29) | θ value | 1.38178e-16 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.420260 (rank : 30) | θ value | 4.59992e-12 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CNTP1_MOUSE
|
||||||
| NC score | 0.418377 (rank : 31) | θ value | 2.28291e-11 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
DDR2_MOUSE
|
||||||
| NC score | 0.161934 (rank : 32) | θ value | 4.59992e-12 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62371 | Gene names | Ddr2, Ntrkr3, Tkt, Tyro10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
DDR2_HUMAN
|
||||||
| NC score | 0.160189 (rank : 33) | θ value | 1.33837e-11 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16832 | Gene names | DDR2, NTRKR3, TKT, TYRO10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
DCBD1_MOUSE
|
||||||
| NC score | 0.157882 (rank : 34) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DDR1_HUMAN
|
||||||
| NC score | 0.148327 (rank : 35) | θ value | 5.08577e-11 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08345, Q14196, Q16562, Q5ST12, Q9UD37 | Gene names | DDR1, CAK, EDDR1, RTK6, TRKE | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (TRK E) (Protein-tyrosine kinase RTK 6) (HGK2) (CD167a antigen). | |||||
|
DDR1_MOUSE
|
||||||
| NC score | 0.141448 (rank : 36) | θ value | 3.29651e-10 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03146 | Gene names | Ddr1, Cak, Eddr1, Mpk6 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (Protein-tyrosine kinase MPK-6) (CD167a antigen). | |||||
|
PCOC2_MOUSE
|
||||||
| NC score | 0.114280 (rank : 37) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.114208 (rank : 38) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.112364 (rank : 39) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PCOC2_HUMAN
|
||||||
| NC score | 0.111528 (rank : 40) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
NETO2_HUMAN
|
||||||
| NC score | 0.101660 (rank : 41) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
CBPE_HUMAN
|
||||||
| NC score | 0.101604 (rank : 42) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16870, Q9UIU9 | Gene names | CPE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPE_MOUSE
|
||||||
| NC score | 0.101487 (rank : 43) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPN_MOUSE
|
||||||
| NC score | 0.100235 (rank : 44) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
CBPN_HUMAN
|
||||||
| NC score | 0.099891 (rank : 45) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15169 | Gene names | CPN1, ACBP | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit) (Lysine carboxypeptidase) (Arginine carboxypeptidase) (Kininase-1) (Serum carboxypeptidase N) (SCPN) (Anaphylatoxin inactivator) (Plasma carboxypeptidase B). | |||||
|
NETO2_MOUSE
|
||||||
| NC score | 0.099674 (rank : 46) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
GLPC_HUMAN
|
||||||
| NC score | 0.099204 (rank : 47) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |

|
|||||
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
NETO1_MOUSE
|
||||||
| NC score | 0.098321 (rank : 48) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.096797 (rank : 49) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.096279 (rank : 50) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
NETO1_HUMAN
|
||||||
| NC score | 0.095723 (rank : 51) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
CBPM_MOUSE
|
||||||
| NC score | 0.091368 (rank : 52) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V42, Q497S5, Q9CYH8 | Gene names | Cpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPM_HUMAN
|
||||||
| NC score | 0.091017 (rank : 53) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14384, Q9H2K9 | Gene names | CPM | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
NRX1A_HUMAN
|
||||||
| NC score | 0.090912 (rank : 54) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
CBPZ_MOUSE
|
||||||
| NC score | 0.090574 (rank : 55) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
NRX1A_MOUSE
|
||||||
| NC score | 0.089870 (rank : 56) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CS84, O88722, Q80Y87, Q8CHE6 | Gene names | Nrxn1, Kiaa0578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
CBPZ_HUMAN
|
||||||
| NC score | 0.089221 (rank : 57) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
PDGFD_HUMAN
|
||||||
| NC score | 0.088712 (rank : 58) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.087792 (rank : 59) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
NRX3A_HUMAN
|
||||||
| NC score | 0.087718 (rank : 60) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4C0, O95378, Q9NS47, Q9P1V3, Q9P1V6, Q9UIE2, Q9UIE3, Q9ULA5, Q9Y486 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-3-alpha precursor (Neurexin III-alpha). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.087610 (rank : 61) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
CBPD_HUMAN
|
||||||
| NC score | 0.087576 (rank : 62) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75976, O15377, Q86SH9, Q86XE6 | Gene names | CPD | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPD_MOUSE
|
||||||
| NC score | 0.087566 (rank : 63) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89001 | Gene names | Cpd | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.087115 (rank : 64) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.082402 (rank : 65) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.081092 (rank : 66) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.079951 (rank : 67) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.077069 (rank : 68) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
BMP1_HUMAN
|
||||||
| NC score | 0.075857 (rank : 69) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| NC score | 0.075747 (rank : 70) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
TLL2_MOUSE
|
||||||
| NC score | 0.075473 (rank : 71) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL1_HUMAN
|
||||||
| NC score | 0.074952 (rank : 72) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TLL2_HUMAN
|
||||||
| NC score | 0.074937 (rank : 73) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
PDGFD_MOUSE
|
||||||
| NC score | 0.074897 (rank : 74) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
TLL1_MOUSE
|
||||||
| NC score | 0.074081 (rank : 75) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
MAMC2_HUMAN
|
||||||
| NC score | 0.068641 (rank : 76) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z304, Q5VW47, Q8WX43, Q96BM4 | Gene names | MAMDC2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.065125 (rank : 77) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.064007 (rank : 78) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
MAMC2_MOUSE
|
||||||
| NC score | 0.063999 (rank : 79) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG85, Q8BM24 | Gene names | Mamdc2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.060786 (rank : 80) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.058237 (rank : 81) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.056301 (rank : 82) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
LRP12_HUMAN
|
||||||
| NC score | 0.056176 (rank : 83) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
GP126_HUMAN
|
||||||
| NC score | 0.054820 (rank : 84) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
UBP2L_MOUSE
|
||||||
| NC score | 0.054557 (rank : 85) | θ value | 0.125558 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
CSMD3_HUMAN
|
||||||
| NC score | 0.053220 (rank : 86) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
SHBG_HUMAN
|
||||||
| NC score | 0.053035 (rank : 87) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04278, P14689, Q16616 | Gene names | SHBG | |||
|
Domain Architecture |

|
|||||
| Description | Sex hormone-binding globulin precursor (SHBG) (Sex steroid-binding protein) (SBP) (Testis-specific androgen-binding protein) (ABP) (Testosterone-estrogen-binding globulin) (Testosterone-estradiol- binding globulin) (TeBG). | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.052666 (rank : 88) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CRUM1_MOUSE
|
||||||
| NC score | 0.052456 (rank : 89) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CSMD1_MOUSE
|
||||||
| NC score | 0.052321 (rank : 90) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CRUM1_HUMAN
|
||||||
| NC score | 0.051716 (rank : 91) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
UBP2L_HUMAN
|
||||||
| NC score | 0.050703 (rank : 92) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.046316 (rank : 93) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.043829 (rank : 94) | θ value | 1.87187e-05 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.040531 (rank : 95) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.039479 (rank : 96) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
TERF2_HUMAN
|
||||||
| NC score | 0.039225 (rank : 97) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TERF2_MOUSE
|
||||||
| NC score | 0.037582 (rank : 98) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.036384 (rank : 99) | θ value | 0.00509761 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
XTP3B_HUMAN
|
||||||
| NC score | 0.036085 (rank : 100) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DZ1, O95901, Q6UWN7, Q9NUY7, Q9UQL4 | Gene names | C2orf30, XTP3TPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XTP3-transactivated gene B protein precursor. | |||||
|
AKAP5_HUMAN
|
||||||
| NC score | 0.035465 (rank : 101) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24588 | Gene names | AKAP5, AKAP79 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 5 (A-kinase anchor protein 79 kDa) (AKAP 79) (cAMP-dependent protein kinase regulatory subunit II high affinity- binding protein) (H21). | |||||
|
XTP3B_MOUSE
|
||||||
| NC score | 0.035043 (rank : 102) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEH8, Q3UZF3, Q8BVN6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XTP3-transactivated gene B protein homolog precursor. | |||||
|
NRIP1_MOUSE
|
||||||
| NC score | 0.033699 (rank : 103) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
TM117_MOUSE
|
||||||
| NC score | 0.033195 (rank : 104) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BH18, Q8C6X8 | Gene names | Tmem117 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 117. | |||||
|
RIF1_HUMAN
|
||||||
| NC score | 0.028172 (rank : 105) | θ value | 0.0961366 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
TERF1_HUMAN
|
||||||
| NC score | 0.025412 (rank : 106) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54274, Q15553, Q93029 | Gene names | TERF1, PIN2, TRF, TRF1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 1 (TTAGGG repeat-binding factor 1) (NIMA-interacting protein 2) (Telomeric protein Pin2/TRF1). | |||||
|
NRIP1_HUMAN
|
||||||
| NC score | 0.023258 (rank : 107) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.020479 (rank : 108) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
TOM70_MOUSE
|
||||||
| NC score | 0.020057 (rank : 109) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.020012 (rank : 110) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.018848 (rank : 111) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.018813 (rank : 112) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.018139 (rank : 113) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
CA026_MOUSE
|
||||||
| NC score | 0.018101 (rank : 114) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.016989 (rank : 115) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.016979 (rank : 116) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.016538 (rank : 117) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.015808 (rank : 118) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
TF3C1_HUMAN
|
||||||
| NC score | 0.015562 (rank : 119) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.015182 (rank : 120) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
TBCD1_HUMAN
|
||||||
| NC score | 0.014642 (rank : 121) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.013908 (rank : 122) | θ value | 0.0563607 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.012817 (rank : 123) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.012592 (rank : 124) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.011965 (rank : 125) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.011904 (rank : 126) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.011304 (rank : 127) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.011259 (rank : 128) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.010617 (rank : 129) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
ICOSL_HUMAN
|
||||||
| NC score | 0.009109 (rank : 130) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75144, Q9HD18, Q9NRQ1 | Gene names | ICOSLG, B7H2, B7RP1, ICOSL, KIAA0653 | |||
|
Domain Architecture |

|
|||||
| Description | ICOS ligand precursor (B7 homolog 2) (B7-H2) (B7-like protein Gl50) (B7-related protein 1) (B7RP-1) (CD275 antigen). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.008905 (rank : 131) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.008881 (rank : 132) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
PLD1_HUMAN
|
||||||
| NC score | 0.008229 (rank : 133) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13393 | Gene names | PLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choline phosphatase 1) (Phosphatidylcholine-hydrolyzing phospholipase D1) (hPLD1). | |||||
|
PLD1_MOUSE
|
||||||
| NC score | 0.008166 (rank : 134) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z280, O35911 | Gene names | Pld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choline phosphatase 1) (Phosphatidylcholine-hydrolyzing phospholipase D1) (mPLD1). | |||||
|
PODXL_MOUSE
|
||||||
| NC score | 0.007367 (rank : 135) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0M4, Q9ESZ1 | Gene names | Podxl, Pclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.007223 (rank : 136) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
TTC4_HUMAN
|
||||||
| NC score | 0.007019 (rank : 137) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.006775 (rank : 138) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.006696 (rank : 139) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
ZHX1_HUMAN
|
||||||
| NC score | 0.006532 (rank : 140) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.006521 (rank : 141) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.005584 (rank : 142) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.005335 (rank : 143) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.005309 (rank : 144) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
RN168_HUMAN
|
||||||
| NC score | 0.004314 (rank : 145) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
GFAP_MOUSE
|
||||||
| NC score | 0.003951 (rank : 146) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
K1C13_HUMAN
|
||||||
| NC score | 0.003802 (rank : 147) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P13646, Q53G54, Q6AZK5, Q8N240 | Gene names | KRT13 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13). | |||||
|
TRPM6_MOUSE
|
||||||
| NC score | 0.003533 (rank : 148) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
KIF3B_MOUSE
|
||||||
| NC score | 0.003522 (rank : 149) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
K1C13_MOUSE
|
||||||
| NC score | 0.003298 (rank : 150) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08730, Q3V176, Q6PAI1 | Gene names | Krt13, Krt1-13 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13) (47 kDa cytokeratin). | |||||
|
PLK4_MOUSE
|
||||||
| NC score | 0.002894 (rank : 151) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64702, Q6PEP6, Q78EG6, Q8R0I5, Q9CVR6, Q9CVU6 | Gene names | Plk4, Sak, Stk18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.002514 (rank : 152) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
SPB12_MOUSE
|
||||||
| NC score | 0.000498 (rank : 153) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7P9, Q6UKZ3 | Gene names | Serpinb12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B12. | |||||
|
ZBT7B_HUMAN
|
||||||
| NC score | -0.004187 (rank : 154) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15156, Q68DR2, Q96EP2 | Gene names | ZBTB7B, ZBTB15, ZFP67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 7B (Zinc finger protein 67 homolog) (Zfp-67) (Zinc finger protein Th-POK) (T-helper-inducing POZ/Krueppel-like factor) (Krueppel-related zinc finger protein cKrox) (hcKrox) (Zinc finger and BTB domain-containing protein 15). | |||||
|
ZN578_HUMAN
|
||||||
| NC score | -0.004489 (rank : 155) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96N58 | Gene names | ZNF578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 578. | |||||