Please be patient as the page loads
|
CPXM2_HUMAN
|
||||||
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CPXM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990065 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989370 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998446 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CBPE_HUMAN
|
||||||
| θ value | 5.84277e-108 (rank : 5) | NC score | 0.858414 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16870, Q9UIU9 | Gene names | CPE | |||
|
Domain Architecture |
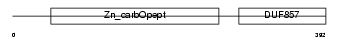
|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPE_MOUSE
|
||||||
| θ value | 4.94618e-107 (rank : 6) | NC score | 0.858124 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |
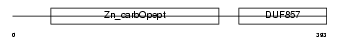
|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPN_HUMAN
|
||||||
| θ value | 4.05562e-101 (rank : 7) | NC score | 0.856071 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15169 | Gene names | CPN1, ACBP | |||
|
Domain Architecture |
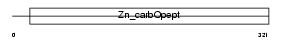
|
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit) (Lysine carboxypeptidase) (Arginine carboxypeptidase) (Kininase-1) (Serum carboxypeptidase N) (SCPN) (Anaphylatoxin inactivator) (Plasma carboxypeptidase B). | |||||
|
CBPN_MOUSE
|
||||||
| θ value | 4.05562e-101 (rank : 8) | NC score | 0.856552 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
CBPZ_HUMAN
|
||||||
| θ value | 2.72037e-97 (rank : 9) | NC score | 0.768763 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPZ_MOUSE
|
||||||
| θ value | 2.54619e-95 (rank : 10) | NC score | 0.783099 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPD_MOUSE
|
||||||
| θ value | 4.81303e-86 (rank : 11) | NC score | 0.816960 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O89001 | Gene names | Cpd | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPD_HUMAN
|
||||||
| θ value | 1.07224e-85 (rank : 12) | NC score | 0.817071 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75976, O15377, Q86SH9, Q86XE6 | Gene names | CPD | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPM_MOUSE
|
||||||
| θ value | 1.83746e-69 (rank : 13) | NC score | 0.832387 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80V42, Q497S5, Q9CYH8 | Gene names | Cpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPM_HUMAN
|
||||||
| θ value | 3.24342e-66 (rank : 14) | NC score | 0.834613 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14384, Q9H2K9 | Gene names | CPM | |||
|
Domain Architecture |
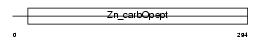
|
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
FA8_MOUSE
|
||||||
| θ value | 4.9032e-22 (rank : 15) | NC score | 0.479324 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA8_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 16) | NC score | 0.478823 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
EDIL3_HUMAN
|
||||||
| θ value | 9.24701e-21 (rank : 17) | NC score | 0.391846 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
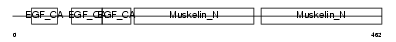
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| θ value | 1.5773e-20 (rank : 18) | NC score | 0.385995 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
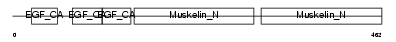
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 19) | NC score | 0.479236 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
MFGM_MOUSE
|
||||||
| θ value | 7.82807e-20 (rank : 20) | NC score | 0.478478 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |
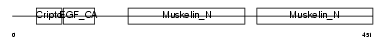
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
NRP1_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 21) | NC score | 0.384078 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
MFGM_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 22) | NC score | 0.550156 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |
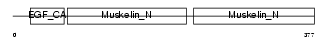
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
NRP2_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 23) | NC score | 0.393915 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
XLRS1_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 24) | NC score | 0.607474 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
NRP2_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 25) | NC score | 0.394504 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
XLRS1_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 26) | NC score | 0.605790 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
DCBD2_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 27) | NC score | 0.454876 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |
|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
NRP1_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 28) | NC score | 0.381707 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 29) | NC score | 0.440620 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
CNTP2_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 30) | NC score | 0.398723 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
DDR2_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 31) | NC score | 0.143686 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62371 | Gene names | Ddr2, Ntrkr3, Tkt, Tyro10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
DDR2_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 32) | NC score | 0.142528 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16832 | Gene names | DDR2, NTRKR3, TKT, TYRO10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
CNTP2_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 33) | NC score | 0.391384 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
DDR1_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 34) | NC score | 0.132991 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08345, Q14196, Q16562, Q5ST12, Q9UD37 | Gene names | DDR1, CAK, EDDR1, RTK6, TRKE | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (TRK E) (Protein-tyrosine kinase RTK 6) (HGK2) (CD167a antigen). | |||||
|
DCBD1_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 35) | NC score | 0.442569 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DDR1_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 36) | NC score | 0.126694 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03146 | Gene names | Ddr1, Cak, Eddr1, Mpk6 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (Protein-tyrosine kinase MPK-6) (CD167a antigen). | |||||
|
CNTP3_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 37) | NC score | 0.387389 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
CNTP1_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 38) | NC score | 0.365134 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 39) | NC score | 0.364828 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CNTP4_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 40) | NC score | 0.374355 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP4_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 41) | NC score | 0.371125 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CBPB2_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 42) | NC score | 0.238818 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JHH6, Q5EBI3, Q9QZF0 | Gene names | Cpb2, Tafi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Carboxypeptidase R) (CPR). | |||||
|
CBPA4_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 43) | NC score | 0.264189 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UI42, Q86UY9 | Gene names | CPA4, CPA3 | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase A4 precursor (EC 3.4.17.-) (Carboxypeptidase A3). | |||||
|
CBPB2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 44) | NC score | 0.196598 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96IY4, Q15114, Q5T9K1, Q5T9K2, Q9P2Y6 | Gene names | CPB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Plasma carboxypeptidase B) (pCPB). | |||||
|
CBPA6_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 45) | NC score | 0.222971 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5U901, Q8BVD0 | Gene names | Cpa6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A6 precursor (EC 3.4.17.1). | |||||
|
CBPA3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 46) | NC score | 0.166159 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15089 | Gene names | Cpa3 | |||
|
Domain Architecture |
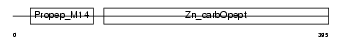
|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
CBPA4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 47) | NC score | 0.223392 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P8K8, Q8BMK6, Q9CTE6 | Gene names | Cpa4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A4 precursor (EC 3.4.17.-). | |||||
|
CBPB1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.135054 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15086, O60834, Q96BQ8 | Gene names | CPB1, CPB, PCPB | |||
|
Domain Architecture |
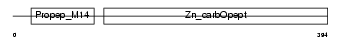
|
|||||
| Description | Carboxypeptidase B precursor (EC 3.4.17.2) (Pancreas-specific protein) (PASP). | |||||
|
CBPA2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.201564 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48052, Q96A12, Q96QN3 | Gene names | CPA2 | |||
|
Domain Architecture |
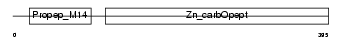
|
|||||
| Description | Carboxypeptidase A2 precursor (EC 3.4.17.15). | |||||
|
CBPO_HUMAN
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.141027 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IVL8, Q2M277, Q7RTW7 | Gene names | CPO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase O precursor (EC 3.4.17.-) (CPO). | |||||
|
CDC20_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.010956 (rank : 93) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12834, Q9BW56, Q9UQI9 | Gene names | CDC20 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 20 homolog (p55CDC). | |||||
|
CDC20_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.010901 (rank : 95) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJ66, Q3TGP1, Q8BPG4, Q99LK3 | Gene names | Cdc20 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 20 homolog (p55CDC) (mmCdc20). | |||||
|
CBPA6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.208505 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4T0, Q8NEX8, Q8TDE8, Q9NRI9 | Gene names | CPA6, CPAH | |||
|
Domain Architecture |
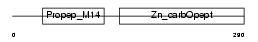
|
|||||
| Description | Carboxypeptidase A6 precursor (EC 3.4.17.1) (Carboxypeptidase B). | |||||
|
CBPA3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.158573 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15088, Q96E94 | Gene names | CPA3 | |||
|
Domain Architecture |
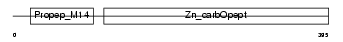
|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | -0.001112 (rank : 112) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.003164 (rank : 106) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.021156 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.006250 (rank : 102) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
AL2S4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.012696 (rank : 92) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.016370 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.016641 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.016902 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.016342 (rank : 89) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.016395 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.016340 (rank : 90) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.010950 (rank : 94) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.004514 (rank : 104) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CHSS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.006620 (rank : 101) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.001798 (rank : 110) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
IL16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.004127 (rank : 105) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.002165 (rank : 109) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
ZBT38_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | -0.004447 (rank : 115) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.005649 (rank : 103) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
ERCC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.007259 (rank : 99) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.002246 (rank : 108) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NOL6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.010464 (rank : 96) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R5K4, Q6PAN8, Q8C6V4, Q8R5K3, Q8VCG0, Q8WTY7 | Gene names | Nol6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.007063 (rank : 100) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | -0.004315 (rank : 114) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.009155 (rank : 98) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.015239 (rank : 91) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
JIP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.001415 (rank : 111) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
MGR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.002440 (rank : 107) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
NOL6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.009826 (rank : 97) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H6R4, Q5T5M3, Q5T5M4, Q7L4G6, Q8N6I0, Q8TEY9, Q8TEZ0, Q8TEZ1, Q9H675 | Gene names | NOL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | -0.001865 (rank : 113) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CBPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.109860 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15085, Q9BS67 | Gene names | CPA1, CPA | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A1 precursor (EC 3.4.17.1). | |||||
|
CBPA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.110693 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TPZ8 | Gene names | Cpa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A1 precursor (EC 3.4.17.1). | |||||
|
CBPA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.135511 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WXQ8, Q86SE2, Q86XM3, Q8NA08 | Gene names | CPA5 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A5 precursor (EC 3.4.17.1). | |||||
|
CBPA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.167539 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R4H4 | Gene names | Cpa5 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A5 precursor (EC 3.4.17.1). | |||||
|
CERU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.092420 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
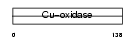
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
CERU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.091480 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.057114 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.056921 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.058185 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.054709 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
DCBD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.111306 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
GLPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.091512 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |
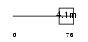
|
|||||
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
HEPH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.091866 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
HEPH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.093155 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
MFRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.073544 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.074938 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NETO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.062405 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.064131 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.066559 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.065314 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NRX1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.078067 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.077106 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CS84, O88722, Q80Y87, Q8CHE6 | Gene names | Nrxn1, Kiaa0578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.066746 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.076518 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4C0, O95378, Q9NS47, Q9P1V3, Q9P1V6, Q9UIE2, Q9UIE3, Q9ULA5, Q9Y486 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |
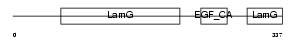
|
|||||
| Description | Neurexin-3-alpha precursor (Neurexin III-alpha). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.074113 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.075475 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.073635 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.075607 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PDGFD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.058426 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.064343 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.064589 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
CPXM2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_MOUSE
|
||||||
| NC score | 0.998446 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM1_HUMAN
|
||||||
| NC score | 0.990065 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM1_MOUSE
|
||||||
| NC score | 0.989370 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CBPE_HUMAN
|
||||||
| NC score | 0.858414 (rank : 5) | θ value | 5.84277e-108 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16870, Q9UIU9 | Gene names | CPE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPE_MOUSE
|
||||||
| NC score | 0.858124 (rank : 6) | θ value | 4.94618e-107 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPN_MOUSE
|
||||||
| NC score | 0.856552 (rank : 7) | θ value | 4.05562e-101 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
CBPN_HUMAN
|
||||||
| NC score | 0.856071 (rank : 8) | θ value | 4.05562e-101 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15169 | Gene names | CPN1, ACBP | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit) (Lysine carboxypeptidase) (Arginine carboxypeptidase) (Kininase-1) (Serum carboxypeptidase N) (SCPN) (Anaphylatoxin inactivator) (Plasma carboxypeptidase B). | |||||
|
CBPM_HUMAN
|
||||||
| NC score | 0.834613 (rank : 9) | θ value | 3.24342e-66 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14384, Q9H2K9 | Gene names | CPM | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPM_MOUSE
|
||||||
| NC score | 0.832387 (rank : 10) | θ value | 1.83746e-69 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80V42, Q497S5, Q9CYH8 | Gene names | Cpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPD_HUMAN
|
||||||
| NC score | 0.817071 (rank : 11) | θ value | 1.07224e-85 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75976, O15377, Q86SH9, Q86XE6 | Gene names | CPD | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPD_MOUSE
|
||||||
| NC score | 0.816960 (rank : 12) | θ value | 4.81303e-86 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O89001 | Gene names | Cpd | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPZ_MOUSE
|
||||||
| NC score | 0.783099 (rank : 13) | θ value | 2.54619e-95 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPZ_HUMAN
|
||||||
| NC score | 0.768763 (rank : 14) | θ value | 2.72037e-97 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
XLRS1_MOUSE
|
||||||
| NC score | 0.607474 (rank : 15) | θ value | 2.7842e-17 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
XLRS1_HUMAN
|
||||||
| NC score | 0.605790 (rank : 16) | θ value | 1.058e-16 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
MFGM_HUMAN
|
||||||
| NC score | 0.550156 (rank : 17) | θ value | 2.7842e-17 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
FA8_MOUSE
|
||||||
| NC score | 0.479324 (rank : 18) | θ value | 4.9032e-22 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.479236 (rank : 19) | θ value | 1.5773e-20 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.478823 (rank : 20) | θ value | 1.86321e-21 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
MFGM_MOUSE
|
||||||
| NC score | 0.478478 (rank : 21) | θ value | 7.82807e-20 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
DCBD2_MOUSE
|
||||||
| NC score | 0.454876 (rank : 22) | θ value | 2.35696e-16 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD1_HUMAN
|
||||||
| NC score | 0.442569 (rank : 23) | θ value | 2.69671e-12 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.440620 (rank : 24) | θ value | 3.07829e-16 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
CNTP2_HUMAN
|
||||||
| NC score | 0.398723 (rank : 25) | θ value | 2.60593e-15 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
NRP2_MOUSE
|
||||||
| NC score | 0.394504 (rank : 26) | θ value | 6.20254e-17 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_HUMAN
|
||||||
| NC score | 0.393915 (rank : 27) | θ value | 2.7842e-17 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
EDIL3_HUMAN
|
||||||
| NC score | 0.391846 (rank : 28) | θ value | 9.24701e-21 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
CNTP2_MOUSE
|
||||||
| NC score | 0.391384 (rank : 29) | θ value | 3.76295e-14 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP3_HUMAN
|
||||||
| NC score | 0.387389 (rank : 30) | θ value | 6.00763e-12 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
EDIL3_MOUSE
|
||||||
| NC score | 0.385995 (rank : 31) | θ value | 1.5773e-20 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
NRP1_HUMAN
|
||||||
| NC score | 0.384078 (rank : 32) | θ value | 5.60996e-18 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
NRP1_MOUSE
|
||||||
| NC score | 0.381707 (rank : 33) | θ value | 2.35696e-16 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
CNTP4_MOUSE
|
||||||
| NC score | 0.374355 (rank : 34) | θ value | 5.08577e-11 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP4_HUMAN
|
||||||
| NC score | 0.371125 (rank : 35) | θ value | 8.67504e-11 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP1_MOUSE
|
||||||
| NC score | 0.365134 (rank : 36) | θ value | 1.74796e-11 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.364828 (rank : 37) | θ value | 2.28291e-11 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CBPA4_HUMAN
|
||||||
| NC score | 0.264189 (rank : 38) | θ value | 0.000602161 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UI42, Q86UY9 | Gene names | CPA4, CPA3 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A4 precursor (EC 3.4.17.-) (Carboxypeptidase A3). | |||||
|
CBPB2_MOUSE
|
||||||
| NC score | 0.238818 (rank : 39) | θ value | 0.000121331 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JHH6, Q5EBI3, Q9QZF0 | Gene names | Cpb2, Tafi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Carboxypeptidase R) (CPR). | |||||
|
CBPA4_MOUSE
|
||||||
| NC score | 0.223392 (rank : 40) | θ value | 0.125558 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P8K8, Q8BMK6, Q9CTE6 | Gene names | Cpa4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A4 precursor (EC 3.4.17.-). | |||||
|
CBPA6_MOUSE
|
||||||
| NC score | 0.222971 (rank : 41) | θ value | 0.0113563 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5U901, Q8BVD0 | Gene names | Cpa6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A6 precursor (EC 3.4.17.1). | |||||
|
CBPA6_HUMAN
|
||||||
| NC score | 0.208505 (rank : 42) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4T0, Q8NEX8, Q8TDE8, Q9NRI9 | Gene names | CPA6, CPAH | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A6 precursor (EC 3.4.17.1) (Carboxypeptidase B). | |||||
|
CBPA2_HUMAN
|
||||||
| NC score | 0.201564 (rank : 43) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48052, Q96A12, Q96QN3 | Gene names | CPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A2 precursor (EC 3.4.17.15). | |||||
|
CBPB2_HUMAN
|
||||||
| NC score | 0.196598 (rank : 44) | θ value | 0.00134147 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96IY4, Q15114, Q5T9K1, Q5T9K2, Q9P2Y6 | Gene names | CPB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Plasma carboxypeptidase B) (pCPB). | |||||
|
CBPA5_MOUSE
|
||||||
| NC score | 0.167539 (rank : 45) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R4H4 | Gene names | Cpa5 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A5 precursor (EC 3.4.17.1). | |||||
|
CBPA3_MOUSE
|
||||||
| NC score | 0.166159 (rank : 46) | θ value | 0.125558 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15089 | Gene names | Cpa3 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
CBPA3_HUMAN
|
||||||
| NC score | 0.158573 (rank : 47) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15088, Q96E94 | Gene names | CPA3 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
DDR2_MOUSE
|
||||||
| NC score | 0.143686 (rank : 48) | θ value | 3.40345e-15 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62371 | Gene names | Ddr2, Ntrkr3, Tkt, Tyro10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
DDR2_HUMAN
|
||||||
| NC score | 0.142528 (rank : 49) | θ value | 2.20605e-14 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16832 | Gene names | DDR2, NTRKR3, TKT, TYRO10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
CBPO_HUMAN
|
||||||
| NC score | 0.141027 (rank : 50) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IVL8, Q2M277, Q7RTW7 | Gene names | CPO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase O precursor (EC 3.4.17.-) (CPO). | |||||
|
CBPA5_HUMAN
|
||||||
| NC score | 0.135511 (rank : 51) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WXQ8, Q86SE2, Q86XM3, Q8NA08 | Gene names | CPA5 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A5 precursor (EC 3.4.17.1). | |||||
|
CBPB1_HUMAN
|
||||||
| NC score | 0.135054 (rank : 52) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15086, O60834, Q96BQ8 | Gene names | CPB1, CPB, PCPB | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase B precursor (EC 3.4.17.2) (Pancreas-specific protein) (PASP). | |||||
|
DDR1_HUMAN
|
||||||
| NC score | 0.132991 (rank : 53) | θ value | 4.16044e-13 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08345, Q14196, Q16562, Q5ST12, Q9UD37 | Gene names | DDR1, CAK, EDDR1, RTK6, TRKE | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (TRK E) (Protein-tyrosine kinase RTK 6) (HGK2) (CD167a antigen). | |||||
|
DDR1_MOUSE
|
||||||
| NC score | 0.126694 (rank : 54) | θ value | 4.59992e-12 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03146 | Gene names | Ddr1, Cak, Eddr1, Mpk6 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (Protein-tyrosine kinase MPK-6) (CD167a antigen). | |||||
|
DCBD1_MOUSE
|
||||||
| NC score | 0.111306 (rank : 55) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
CBPA1_MOUSE
|
||||||
| NC score | 0.110693 (rank : 56) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TPZ8 | Gene names | Cpa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A1 precursor (EC 3.4.17.1). | |||||
|
CBPA1_HUMAN
|
||||||
| NC score | 0.109860 (rank : 57) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15085, Q9BS67 | Gene names | CPA1, CPA | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A1 precursor (EC 3.4.17.1). | |||||
|
HEPH_MOUSE
|
||||||
| NC score | 0.093155 (rank : 58) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
CERU_HUMAN
|
||||||
| NC score | 0.092420 (rank : 59) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
HEPH_HUMAN
|
||||||
| NC score | 0.091866 (rank : 60) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
GLPC_HUMAN
|
||||||
| NC score | 0.091512 (rank : 61) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |

|
|||||
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
CERU_MOUSE
|
||||||
| NC score | 0.091480 (rank : 62) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
NRX1A_HUMAN
|
||||||
| NC score | 0.078067 (rank : 63) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX1A_MOUSE
|
||||||
| NC score | 0.077106 (rank : 64) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CS84, O88722, Q80Y87, Q8CHE6 | Gene names | Nrxn1, Kiaa0578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX3A_HUMAN
|
||||||
| NC score | 0.076518 (rank : 65) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4C0, O95378, Q9NS47, Q9P1V3, Q9P1V6, Q9UIE2, Q9UIE3, Q9ULA5, Q9Y486 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-3-alpha precursor (Neurexin III-alpha). | |||||
|
PCOC2_MOUSE
|
||||||
| NC score | 0.075607 (rank : 66) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.075475 (rank : 67) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.074938 (rank : 68) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.074113 (rank : 69) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PCOC2_HUMAN
|
||||||
| NC score | 0.073635 (rank : 70) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.073544 (rank : 71) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.066746 (rank : 72) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NETO2_HUMAN
|
||||||
| NC score | 0.066559 (rank : 73) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| NC score | 0.065314 (rank : 74) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.064589 (rank : 75) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.064343 (rank : 76) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
NETO1_MOUSE
|
||||||
| NC score | 0.064131 (rank : 77) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_HUMAN
|
||||||
| NC score | 0.062405 (rank : 78) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
PDGFD_HUMAN
|
||||||
| NC score | 0.058426 (rank : 79) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.058185 (rank : 80) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.057114 (rank : 81) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.056921 (rank : 82) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.054709 (rank : 83) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.021156 (rank : 84) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.016902 (rank : 85) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.016641 (rank : 86) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.016395 (rank : 87) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.016370 (rank : 88) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.016342 (rank : 89) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.016340 (rank : 90) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.015239 (rank : 91) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
AL2S4_MOUSE
|
||||||
| NC score | 0.012696 (rank : 92) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
CDC20_HUMAN
|
||||||
| NC score | 0.010956 (rank : 93) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12834, Q9BW56, Q9UQI9 | Gene names | CDC20 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 20 homolog (p55CDC). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.010950 (rank : 94) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
CDC20_MOUSE
|
||||||
| NC score | 0.010901 (rank : 95) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJ66, Q3TGP1, Q8BPG4, Q99LK3 | Gene names | Cdc20 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 20 homolog (p55CDC) (mmCdc20). | |||||
|
NOL6_MOUSE
|
||||||
| NC score | 0.010464 (rank : 96) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R5K4, Q6PAN8, Q8C6V4, Q8R5K3, Q8VCG0, Q8WTY7 | Gene names | Nol6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
NOL6_HUMAN
|
||||||
| NC score | 0.009826 (rank : 97) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H6R4, Q5T5M3, Q5T5M4, Q7L4G6, Q8N6I0, Q8TEY9, Q8TEZ0, Q8TEZ1, Q9H675 | Gene names | NOL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.009155 (rank : 98) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
ERCC4_MOUSE
|
||||||
| NC score | 0.007259 (rank : 99) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.007063 (rank : 100) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
CHSS2_MOUSE
|
||||||
| NC score | 0.006620 (rank : 101) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.006250 (rank : 102) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.005649 (rank : 103) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.004514 (rank : 104) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
IL16_HUMAN
|
||||||
| NC score | 0.004127 (rank : 105) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.003164 (rank : 106) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MGR3_HUMAN
|
||||||
| NC score | 0.002440 (rank : 107) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.002246 (rank : 108) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.002165 (rank : 109) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.001798 (rank : 110) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
JIP4_MOUSE
|
||||||
| NC score | 0.001415 (rank : 111) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
NFH_MOUSE
|
||||||
| NC score | -0.001112 (rank : 112) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | -0.001865 (rank : 113) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | -0.004315 (rank : 114) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZBT38_HUMAN
|
||||||
| NC score | -0.004447 (rank : 115) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||