Please be patient as the page loads
|
CBPD_HUMAN
|
||||||
| SwissProt Accessions | O75976, O15377, Q86SH9, Q86XE6 | Gene names | CPD | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CBPD_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75976, O15377, Q86SH9, Q86XE6 | Gene names | CPD | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPD_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998753 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O89001 | Gene names | Cpd | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPE_HUMAN
|
||||||
| θ value | 4.47364e-108 (rank : 3) | NC score | 0.953565 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16870, Q9UIU9 | Gene names | CPE | |||
|
Domain Architecture |
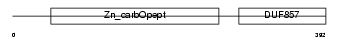
|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPE_MOUSE
|
||||||
| θ value | 9.96626e-108 (rank : 4) | NC score | 0.953676 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |
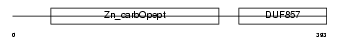
|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPN_HUMAN
|
||||||
| θ value | 9.32813e-106 (rank : 5) | NC score | 0.956010 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15169 | Gene names | CPN1, ACBP | |||
|
Domain Architecture |
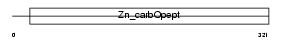
|
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit) (Lysine carboxypeptidase) (Arginine carboxypeptidase) (Kininase-1) (Serum carboxypeptidase N) (SCPN) (Anaphylatoxin inactivator) (Plasma carboxypeptidase B). | |||||
|
CBPN_MOUSE
|
||||||
| θ value | 1.64657e-102 (rank : 6) | NC score | 0.954146 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
CBPM_MOUSE
|
||||||
| θ value | 3.66817e-102 (rank : 7) | NC score | 0.961945 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80V42, Q497S5, Q9CYH8 | Gene names | Cpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPM_HUMAN
|
||||||
| θ value | 8.17181e-102 (rank : 8) | NC score | 0.958971 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14384, Q9H2K9 | Gene names | CPM | |||
|
Domain Architecture |
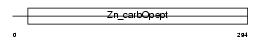
|
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CPXM1_HUMAN
|
||||||
| θ value | 2.38316e-93 (rank : 9) | NC score | 0.844918 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM1_MOUSE
|
||||||
| θ value | 9.056e-93 (rank : 10) | NC score | 0.848888 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM2_HUMAN
|
||||||
| θ value | 1.07224e-85 (rank : 11) | NC score | 0.817071 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_MOUSE
|
||||||
| θ value | 1.07224e-85 (rank : 12) | NC score | 0.815665 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CBPZ_HUMAN
|
||||||
| θ value | 3.44927e-84 (rank : 13) | NC score | 0.851055 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPZ_MOUSE
|
||||||
| θ value | 1.31072e-83 (rank : 14) | NC score | 0.870277 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPA4_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 15) | NC score | 0.459626 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UI42, Q86UY9 | Gene names | CPA4, CPA3 | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase A4 precursor (EC 3.4.17.-) (Carboxypeptidase A3). | |||||
|
CBPA2_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 16) | NC score | 0.397852 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48052, Q96A12, Q96QN3 | Gene names | CPA2 | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase A2 precursor (EC 3.4.17.15). | |||||
|
CBPA4_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 17) | NC score | 0.418196 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P8K8, Q8BMK6, Q9CTE6 | Gene names | Cpa4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A4 precursor (EC 3.4.17.-). | |||||
|
CBPB2_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 18) | NC score | 0.415534 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JHH6, Q5EBI3, Q9QZF0 | Gene names | Cpb2, Tafi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Carboxypeptidase R) (CPR). | |||||
|
CBPB2_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 19) | NC score | 0.373848 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96IY4, Q15114, Q5T9K1, Q5T9K2, Q9P2Y6 | Gene names | CPB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Plasma carboxypeptidase B) (pCPB). | |||||
|
CBPA1_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 20) | NC score | 0.303375 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TPZ8 | Gene names | Cpa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A1 precursor (EC 3.4.17.1). | |||||
|
CBPA6_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 21) | NC score | 0.401572 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5U901, Q8BVD0 | Gene names | Cpa6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A6 precursor (EC 3.4.17.1). | |||||
|
CBPA1_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 22) | NC score | 0.300270 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15085, Q9BS67 | Gene names | CPA1, CPA | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A1 precursor (EC 3.4.17.1). | |||||
|
CBPA5_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 23) | NC score | 0.365547 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8R4H4 | Gene names | Cpa5 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A5 precursor (EC 3.4.17.1). | |||||
|
CBPA6_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 24) | NC score | 0.387960 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N4T0, Q8NEX8, Q8TDE8, Q9NRI9 | Gene names | CPA6, CPAH | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase A6 precursor (EC 3.4.17.1) (Carboxypeptidase B). | |||||
|
CBPA3_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 25) | NC score | 0.337557 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15088, Q96E94 | Gene names | CPA3 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
CBPO_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 26) | NC score | 0.319007 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IVL8, Q2M277, Q7RTW7 | Gene names | CPO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase O precursor (EC 3.4.17.-) (CPO). | |||||
|
CBPA5_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 27) | NC score | 0.329682 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXQ8, Q86SE2, Q86XM3, Q8NA08 | Gene names | CPA5 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A5 precursor (EC 3.4.17.1). | |||||
|
CBPB1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 28) | NC score | 0.310621 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15086, O60834, Q96BQ8 | Gene names | CPB1, CPB, PCPB | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase B precursor (EC 3.4.17.2) (Pancreas-specific protein) (PASP). | |||||
|
CBPA3_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 29) | NC score | 0.338026 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15089 | Gene names | Cpa3 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.022599 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
ZBED1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.050325 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O96006, Q96BY4 | Gene names | ZBED1, ALTE, DREF, KIAA0785, TRAMP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 1 (dREF homolog) (Putative Ac-like transposable element). | |||||
|
NLGN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.016615 (rank : 58) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.016608 (rank : 59) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |
|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.007128 (rank : 70) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
FLNC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.007233 (rank : 69) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14315, O95303, Q07985, Q9NS12, Q9NYE5, Q9UMR8, Q9Y503 | Gene names | FLNC, ABPL, FLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-C (Gamma-filamin) (Filamin-2) (Protein FLNc) (Actin-binding- like protein) (ABP-L) (ABP-280-like protein). | |||||
|
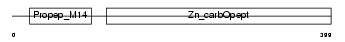
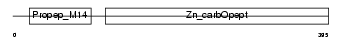
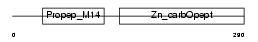
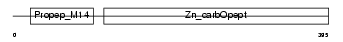
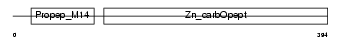
PROP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.012206 (rank : 65) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
Domain Architecture |
|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
HA2D_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.016581 (rank : 61) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04228 | Gene names | H2-Aa | |||
|
Domain Architecture |
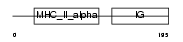
|
|||||
| Description | H-2 class II histocompatibility antigen, A-D alpha chain precursor. | |||||
|
HA2F_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.016621 (rank : 57) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14435 | Gene names | H2-Aa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | H-2 class II histocompatibility antigen, A-F alpha chain. | |||||
|
HA2R_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.016601 (rank : 60) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14436 | Gene names | ||||
|
Domain Architecture |
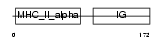
|
|||||
| Description | H-2 class II histocompatibility antigen, A-R alpha chain. | |||||
|
HA2S_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.016714 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14437 | Gene names | ||||
|
Domain Architecture |
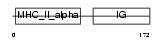
|
|||||
| Description | H-2 class II histocompatibility antigen, A-S alpha chain. | |||||
|
SOX4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.005476 (rank : 72) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
TCPE_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.011201 (rank : 66) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P80316 | Gene names | Cct5, Ccte | |||
|
Domain Architecture |

|
|||||
| Description | T-complex protein 1 subunit epsilon (TCP-1-epsilon) (CCT-epsilon). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.008888 (rank : 67) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CML1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.001038 (rank : 75) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99788, O75748, Q99789 | Gene names | CMKLR1, CHEMR23, DEZ | |||
|
Domain Architecture |
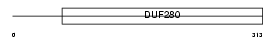
|
|||||
| Description | Chemokine receptor-like 1 (G-protein coupled receptor DEZ) (G-protein coupled receptor ChemR23). | |||||
|
KI18A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.003054 (rank : 74) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WD7, Q3TP77, Q8BLL1 | Gene names | Kif18a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
HA2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.015303 (rank : 62) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14434, O78195 | Gene names | H2-Aa | |||
|
Domain Architecture |
|
|||||
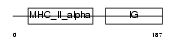
| Description | H-2 class II histocompatibility antigen, A-B alpha chain precursor (IAalpha). | |||||
|
HA2K_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.015109 (rank : 63) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P01910 | Gene names | H2-Aa | |||
|
Domain Architecture |
|
|||||
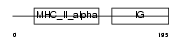
| Description | H-2 class II histocompatibility antigen, A-K alpha chain precursor. | |||||
|
K0423_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.012331 (rank : 64) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
TCPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.007102 (rank : 71) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48643 | Gene names | CCT5, CCTE, KIAA0098 | |||
|
Domain Architecture |

|
|||||
| Description | T-complex protein 1 subunit epsilon (TCP-1-epsilon) (CCT-epsilon). | |||||
|
CND3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.021081 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.007559 (rank : 68) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | -0.000068 (rank : 76) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NCK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.003266 (rank : 73) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |
|
|||||
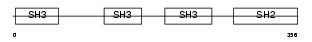
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.055393 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CNTP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.056875 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
CNTP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.066383 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.065579 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.060593 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
CNTP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.051794 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.053559 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
DCBD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.082218 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.081075 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
DCBD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.085819 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |
|
|||||
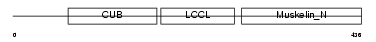
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
EDIL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.074271 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
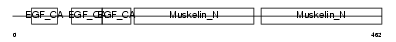
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.072857 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
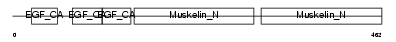
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
FA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.087171 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
FA8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.086468 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
FA8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.087576 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
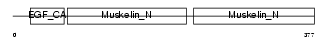
MFGM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.103659 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
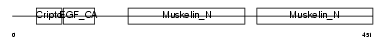
MFGM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.091306 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
NRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.063382 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
NRP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.059744 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
NRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.068765 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.068314 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
XLRS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.139103 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
XLRS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.140682 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
CBPD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75976, O15377, Q86SH9, Q86XE6 | Gene names | CPD | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPD_MOUSE
|
||||||
| NC score | 0.998753 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O89001 | Gene names | Cpd | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPM_MOUSE
|
||||||
| NC score | 0.961945 (rank : 3) | θ value | 3.66817e-102 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80V42, Q497S5, Q9CYH8 | Gene names | Cpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPM_HUMAN
|
||||||
| NC score | 0.958971 (rank : 4) | θ value | 8.17181e-102 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14384, Q9H2K9 | Gene names | CPM | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPN_HUMAN
|
||||||
| NC score | 0.956010 (rank : 5) | θ value | 9.32813e-106 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15169 | Gene names | CPN1, ACBP | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit) (Lysine carboxypeptidase) (Arginine carboxypeptidase) (Kininase-1) (Serum carboxypeptidase N) (SCPN) (Anaphylatoxin inactivator) (Plasma carboxypeptidase B). | |||||
|
CBPN_MOUSE
|
||||||
| NC score | 0.954146 (rank : 6) | θ value | 1.64657e-102 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
CBPE_MOUSE
|
||||||
| NC score | 0.953676 (rank : 7) | θ value | 9.96626e-108 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPE_HUMAN
|
||||||
| NC score | 0.953565 (rank : 8) | θ value | 4.47364e-108 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16870, Q9UIU9 | Gene names | CPE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPZ_MOUSE
|
||||||
| NC score | 0.870277 (rank : 9) | θ value | 1.31072e-83 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPZ_HUMAN
|
||||||
| NC score | 0.851055 (rank : 10) | θ value | 3.44927e-84 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CPXM1_MOUSE
|
||||||
| NC score | 0.848888 (rank : 11) | θ value | 9.056e-93 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM1_HUMAN
|
||||||
| NC score | 0.844918 (rank : 12) | θ value | 2.38316e-93 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM2_HUMAN
|
||||||
| NC score | 0.817071 (rank : 13) | θ value | 1.07224e-85 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_MOUSE
|
||||||
| NC score | 0.815665 (rank : 14) | θ value | 1.07224e-85 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CBPA4_HUMAN
|
||||||
| NC score | 0.459626 (rank : 15) | θ value | 5.43371e-13 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UI42, Q86UY9 | Gene names | CPA4, CPA3 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A4 precursor (EC 3.4.17.-) (Carboxypeptidase A3). | |||||
|
CBPA4_MOUSE
|
||||||
| NC score | 0.418196 (rank : 16) | θ value | 2.13673e-09 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P8K8, Q8BMK6, Q9CTE6 | Gene names | Cpa4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A4 precursor (EC 3.4.17.-). | |||||
|
CBPB2_MOUSE
|
||||||
| NC score | 0.415534 (rank : 17) | θ value | 1.38499e-08 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JHH6, Q5EBI3, Q9QZF0 | Gene names | Cpb2, Tafi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Carboxypeptidase R) (CPR). | |||||
|
CBPA6_MOUSE
|
||||||
| NC score | 0.401572 (rank : 18) | θ value | 1.53129e-07 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5U901, Q8BVD0 | Gene names | Cpa6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A6 precursor (EC 3.4.17.1). | |||||
|
CBPA2_HUMAN
|
||||||
| NC score | 0.397852 (rank : 19) | θ value | 4.30538e-10 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48052, Q96A12, Q96QN3 | Gene names | CPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A2 precursor (EC 3.4.17.15). | |||||
|
CBPA6_HUMAN
|
||||||
| NC score | 0.387960 (rank : 20) | θ value | 1.09739e-05 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N4T0, Q8NEX8, Q8TDE8, Q9NRI9 | Gene names | CPA6, CPAH | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A6 precursor (EC 3.4.17.1) (Carboxypeptidase B). | |||||
|
CBPB2_HUMAN
|
||||||
| NC score | 0.373848 (rank : 21) | θ value | 1.80886e-08 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96IY4, Q15114, Q5T9K1, Q5T9K2, Q9P2Y6 | Gene names | CPB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Plasma carboxypeptidase B) (pCPB). | |||||
|
CBPA5_MOUSE
|
||||||
| NC score | 0.365547 (rank : 22) | θ value | 9.92553e-07 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8R4H4 | Gene names | Cpa5 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A5 precursor (EC 3.4.17.1). | |||||
|
CBPA3_MOUSE
|
||||||
| NC score | 0.338026 (rank : 23) | θ value | 0.000158464 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15089 | Gene names | Cpa3 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
CBPA3_HUMAN
|
||||||
| NC score | 0.337557 (rank : 24) | θ value | 1.43324e-05 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15088, Q96E94 | Gene names | CPA3 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
CBPA5_HUMAN
|
||||||
| NC score | 0.329682 (rank : 25) | θ value | 2.44474e-05 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXQ8, Q86SE2, Q86XM3, Q8NA08 | Gene names | CPA5 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A5 precursor (EC 3.4.17.1). | |||||
|
CBPO_HUMAN
|
||||||
| NC score | 0.319007 (rank : 26) | θ value | 1.87187e-05 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IVL8, Q2M277, Q7RTW7 | Gene names | CPO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase O precursor (EC 3.4.17.-) (CPO). | |||||
|
CBPB1_HUMAN
|
||||||
| NC score | 0.310621 (rank : 27) | θ value | 0.000121331 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15086, O60834, Q96BQ8 | Gene names | CPB1, CPB, PCPB | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase B precursor (EC 3.4.17.2) (Pancreas-specific protein) (PASP). | |||||
|
CBPA1_MOUSE
|
||||||
| NC score | 0.303375 (rank : 28) | θ value | 1.53129e-07 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TPZ8 | Gene names | Cpa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase A1 precursor (EC 3.4.17.1). | |||||
|
CBPA1_HUMAN
|
||||||
| NC score | 0.300270 (rank : 29) | θ value | 5.81887e-07 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15085, Q9BS67 | Gene names | CPA1, CPA | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A1 precursor (EC 3.4.17.1). | |||||
|
XLRS1_MOUSE
|
||||||
| NC score | 0.140682 (rank : 30) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
XLRS1_HUMAN
|
||||||
| NC score | 0.139103 (rank : 31) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
MFGM_HUMAN
|
||||||
| NC score | 0.103659 (rank : 32) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
MFGM_MOUSE
|
||||||
| NC score | 0.091306 (rank : 33) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
FA8_MOUSE
|
||||||
| NC score | 0.087576 (rank : 34) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.087171 (rank : 35) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.086468 (rank : 36) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
DCBD2_MOUSE
|
||||||
| NC score | 0.085819 (rank : 37) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD1_HUMAN
|
||||||
| NC score | 0.082218 (rank : 38) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.081075 (rank : 39) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
EDIL3_HUMAN
|
||||||
| NC score | 0.074271 (rank : 40) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| NC score | 0.072857 (rank : 41) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
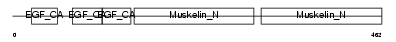
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
NRP2_HUMAN
|
||||||
| NC score | 0.068765 (rank : 42) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_MOUSE
|
||||||
| NC score | 0.068314 (rank : 43) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
CNTP2_HUMAN
|
||||||
| NC score | 0.066383 (rank : 44) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP2_MOUSE
|
||||||
| NC score | 0.065579 (rank : 45) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
NRP1_HUMAN
|
||||||
| NC score | 0.063382 (rank : 46) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
CNTP3_HUMAN
|
||||||
| NC score | 0.060593 (rank : 47) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
NRP1_MOUSE
|
||||||
| NC score | 0.059744 (rank : 48) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
CNTP1_MOUSE
|
||||||
| NC score | 0.056875 (rank : 49) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.055393 (rank : 50) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CNTP4_MOUSE
|
||||||
| NC score | 0.053559 (rank : 51) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP4_HUMAN
|
||||||
| NC score | 0.051794 (rank : 52) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
ZBED1_HUMAN
|
||||||
| NC score | 0.050325 (rank : 53) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O96006, Q96BY4 | Gene names | ZBED1, ALTE, DREF, KIAA0785, TRAMP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 1 (dREF homolog) (Putative Ac-like transposable element). | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.022599 (rank : 54) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
CND3_HUMAN
|
||||||
| NC score | 0.021081 (rank : 55) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
HA2S_MOUSE
|
||||||
| NC score | 0.016714 (rank : 56) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14437 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen, A-S alpha chain. | |||||
|
HA2F_MOUSE
|
||||||
| NC score | 0.016621 (rank : 57) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14435 | Gene names | H2-Aa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | H-2 class II histocompatibility antigen, A-F alpha chain. | |||||
|
NLGN1_HUMAN
|
||||||
| NC score | 0.016615 (rank : 58) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_MOUSE
|
||||||
| NC score | 0.016608 (rank : 59) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
HA2R_MOUSE
|
||||||
| NC score | 0.016601 (rank : 60) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14436 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen, A-R alpha chain. | |||||
|
HA2D_MOUSE
|
||||||
| NC score | 0.016581 (rank : 61) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04228 | Gene names | H2-Aa | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen, A-D alpha chain precursor. | |||||
|
HA2B_MOUSE
|
||||||
| NC score | 0.015303 (rank : 62) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14434, O78195 | Gene names | H2-Aa | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen, A-B alpha chain precursor (IAalpha). | |||||
|
HA2K_MOUSE
|
||||||
| NC score | 0.015109 (rank : 63) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P01910 | Gene names | H2-Aa | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen, A-K alpha chain precursor. | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.012331 (rank : 64) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
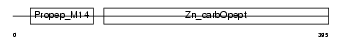
PROP_HUMAN
|
||||||
| NC score | 0.012206 (rank : 65) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
Domain Architecture |

|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
TCPE_MOUSE
|
||||||
| NC score | 0.011201 (rank : 66) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P80316 | Gene names | Cct5, Ccte | |||
|
Domain Architecture |

|
|||||
| Description | T-complex protein 1 subunit epsilon (TCP-1-epsilon) (CCT-epsilon). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.008888 (rank : 67) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.007559 (rank : 68) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
FLNC_HUMAN
|
||||||
| NC score | 0.007233 (rank : 69) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14315, O95303, Q07985, Q9NS12, Q9NYE5, Q9UMR8, Q9Y503 | Gene names | FLNC, ABPL, FLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-C (Gamma-filamin) (Filamin-2) (Protein FLNc) (Actin-binding- like protein) (ABP-L) (ABP-280-like protein). | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.007128 (rank : 70) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
TCPE_HUMAN
|
||||||
| NC score | 0.007102 (rank : 71) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48643 | Gene names | CCT5, CCTE, KIAA0098 | |||
|
Domain Architecture |

|
|||||
| Description | T-complex protein 1 subunit epsilon (TCP-1-epsilon) (CCT-epsilon). | |||||
|
SOX4_HUMAN
|
||||||
| NC score | 0.005476 (rank : 72) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
NCK2_HUMAN
|
||||||
| NC score | 0.003266 (rank : 73) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
KI18A_MOUSE
|
||||||
| NC score | 0.003054 (rank : 74) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WD7, Q3TP77, Q8BLL1 | Gene names | Kif18a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
CML1_HUMAN
|
||||||
| NC score | 0.001038 (rank : 75) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99788, O75748, Q99789 | Gene names | CMKLR1, CHEMR23, DEZ | |||
|
Domain Architecture |

|
|||||
| Description | Chemokine receptor-like 1 (G-protein coupled receptor DEZ) (G-protein coupled receptor ChemR23). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | -0.000068 (rank : 76) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||