Please be patient as the page loads
|
PPIL2_MOUSE
|
||||||
| SwissProt Accessions | Q9D787, Q542A2, Q9CZL1 | Gene names | Ppil2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PPIL2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999322 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13356, Q13357, Q8TAH2, Q9BWR8 | Gene names | PPIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
PPIL2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9D787, Q542A2, Q9CZL1 | Gene names | Ppil2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
PPIL3_HUMAN
|
||||||
| θ value | 7.51208e-47 (rank : 3) | NC score | 0.956556 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H2H8, Q86WF9, Q96IA9, Q9BXZ1 | Gene names | PPIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 3 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL3) (Cyclophilin J) (CyPJ). | |||||
|
PPIL3_MOUSE
|
||||||
| θ value | 4.86918e-46 (rank : 4) | NC score | 0.956371 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D6L8, Q812D2, Q9CY23 | Gene names | Ppil3, Cyp10l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 3 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL3) (CYP10L). | |||||
|
PPWD1_HUMAN
|
||||||
| θ value | 1.08726e-37 (rank : 5) | NC score | 0.868998 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96BP3, Q15002, Q7KZ89 | Gene names | PPWD1, KIAA0073 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
PPWD1_MOUSE
|
||||||
| θ value | 7.04741e-37 (rank : 6) | NC score | 0.869499 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CEC6 | Gene names | Ppwd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
PPIL1_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 7) | NC score | 0.932733 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y3C6, O15001 | Gene names | PPIL1, CYPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 1 (EC 5.2.1.8) (PPIase) (Rotamase). | |||||
|
PPIL1_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 8) | NC score | 0.932799 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D0W5, Q3UA03 | Gene names | Ppil1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 1 (EC 5.2.1.8) (PPIase) (Rotamase). | |||||
|
PPIH_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 9) | NC score | 0.899763 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43447 | Gene names | PPIH, CYP20, CYPH | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H) (U-snRNP-associated cyclophilin SnuCyp-20) (USA-CYP) (Small nuclear ribonucleoprotein particle-specific cyclophilin H) (CypH). | |||||
|
PPIE_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 10) | NC score | 0.762372 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIE_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 11) | NC score | 0.762631 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPID_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 12) | NC score | 0.811108 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
PPIF_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 13) | NC score | 0.888536 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30405, Q5W131 | Gene names | PPIF, CYP3 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPID_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 14) | NC score | 0.804343 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPIH_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 15) | NC score | 0.896086 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D868, Q9CQU7 | Gene names | Ppih | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 16) | NC score | 0.667723 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
PPIA_HUMAN
|
||||||
| θ value | 1.80048e-24 (rank : 17) | NC score | 0.890100 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P62937, P05092, Q6IBU5, Q96IX3, Q9BRU4, Q9BTY9, Q9UC61 | Gene names | PPIA, CYPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein). | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 1.80048e-24 (rank : 18) | NC score | 0.660428 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
PPIA_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 19) | NC score | 0.889872 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17742, Q9CWJ5, Q9R137 | Gene names | Ppia | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein) (SP18). | |||||
|
PPIL4_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 20) | NC score | 0.722707 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
PPIL4_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 21) | NC score | 0.726361 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
PPIC_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 22) | NC score | 0.890574 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30412 | Gene names | Ppic, Cypc | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPIF_MOUSE
|
||||||
| θ value | 1.5242e-23 (rank : 23) | NC score | 0.888590 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KR7 | Gene names | Ppif | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 24) | NC score | 0.768926 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 1.99067e-23 (rank : 25) | NC score | 0.806321 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 7.56453e-23 (rank : 26) | NC score | 0.750702 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
PPIB_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 27) | NC score | 0.875784 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24369 | Gene names | Ppib | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIB_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 28) | NC score | 0.874482 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23284, Q9BVK5 | Gene names | PPIB, CYPB | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIC_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 29) | NC score | 0.878582 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P45877 | Gene names | PPIC, CYPC | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
TF3C1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.044892 (rank : 46) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
TTC6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.039675 (rank : 47) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
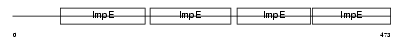
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.005917 (rank : 54) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
SRPK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.007792 (rank : 53) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78362, O75220, O75221, Q6NUL0, Q6V1X2, Q8IYQ3 | Gene names | SRPK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
TF3C1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.035333 (rank : 48) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K284, Q8CAL9 | Gene names | Gtf3c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.008527 (rank : 51) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
MED12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.012197 (rank : 49) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.008343 (rank : 52) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
AN32A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.004582 (rank : 56) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.005293 (rank : 55) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
K1267_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.009596 (rank : 50) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3B3, Q6AW85, Q8IYH1, Q9NTE7, Q9UFT0, Q9ULF3 | Gene names | KIAA1267 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1267. | |||||
|
MMP16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.001705 (rank : 57) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
PRD15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | -0.003662 (rank : 58) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.051913 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
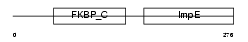
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
FUSIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.062002 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |
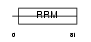
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.062002 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |
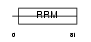
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
NU153_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.089088 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
RANG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.107608 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43487 | Gene names | RANBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RanBP1). | |||||
|
RANG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.108385 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34022, P34023, Q3TL81, Q9D894, Q9DCA3 | Gene names | Ranbp1, Htf9-a, Htf9a | |||
|
Domain Architecture |
|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RANBP1) (HpaII tiny fragments locus 9a protein). | |||||
|
RBP22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.110859 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RBP26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.111479 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
RGPD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.111256 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
SFRS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.052110 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.052110 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
SRR35_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.056964 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |
|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
TRA2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.054547 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |
|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
TRA2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.052680 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PFR5 | Gene names | Tra2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
TRA2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.051890 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.051890 (rank : 45) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
PPIL2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9D787, Q542A2, Q9CZL1 | Gene names | Ppil2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
PPIL2_HUMAN
|
||||||
| NC score | 0.999322 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13356, Q13357, Q8TAH2, Q9BWR8 | Gene names | PPIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
PPIL3_HUMAN
|
||||||
| NC score | 0.956556 (rank : 3) | θ value | 7.51208e-47 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H2H8, Q86WF9, Q96IA9, Q9BXZ1 | Gene names | PPIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 3 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL3) (Cyclophilin J) (CyPJ). | |||||
|
PPIL3_MOUSE
|
||||||
| NC score | 0.956371 (rank : 4) | θ value | 4.86918e-46 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D6L8, Q812D2, Q9CY23 | Gene names | Ppil3, Cyp10l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 3 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL3) (CYP10L). | |||||
|
PPIL1_MOUSE
|
||||||
| NC score | 0.932799 (rank : 5) | θ value | 3.74554e-30 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D0W5, Q3UA03 | Gene names | Ppil1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 1 (EC 5.2.1.8) (PPIase) (Rotamase). | |||||
|
PPIL1_HUMAN
|
||||||
| NC score | 0.932733 (rank : 6) | θ value | 3.74554e-30 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y3C6, O15001 | Gene names | PPIL1, CYPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 1 (EC 5.2.1.8) (PPIase) (Rotamase). | |||||
|
PPIH_HUMAN
|
||||||
| NC score | 0.899763 (rank : 7) | θ value | 2.27234e-27 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43447 | Gene names | PPIH, CYP20, CYPH | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H) (U-snRNP-associated cyclophilin SnuCyp-20) (USA-CYP) (Small nuclear ribonucleoprotein particle-specific cyclophilin H) (CypH). | |||||
|
PPIH_MOUSE
|
||||||
| NC score | 0.896086 (rank : 8) | θ value | 1.37858e-24 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D868, Q9CQU7 | Gene names | Ppih | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H). | |||||
|
PPIC_MOUSE
|
||||||
| NC score | 0.890574 (rank : 9) | θ value | 8.93572e-24 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30412 | Gene names | Ppic, Cypc | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPIA_HUMAN
|
||||||
| NC score | 0.890100 (rank : 10) | θ value | 1.80048e-24 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P62937, P05092, Q6IBU5, Q96IX3, Q9BRU4, Q9BTY9, Q9UC61 | Gene names | PPIA, CYPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein). | |||||
|
PPIA_MOUSE
|
||||||
| NC score | 0.889872 (rank : 11) | θ value | 3.07116e-24 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17742, Q9CWJ5, Q9R137 | Gene names | Ppia | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein) (SP18). | |||||
|
PPIF_MOUSE
|
||||||
| NC score | 0.888590 (rank : 12) | θ value | 1.5242e-23 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KR7 | Gene names | Ppif | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPIF_HUMAN
|
||||||
| NC score | 0.888536 (rank : 13) | θ value | 6.18819e-25 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30405, Q5W131 | Gene names | PPIF, CYP3 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPIC_HUMAN
|
||||||
| NC score | 0.878582 (rank : 14) | θ value | 6.20254e-17 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P45877 | Gene names | PPIC, CYPC | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPIB_MOUSE
|
||||||
| NC score | 0.875784 (rank : 15) | θ value | 4.74913e-17 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24369 | Gene names | Ppib | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIB_HUMAN
|
||||||
| NC score | 0.874482 (rank : 16) | θ value | 6.20254e-17 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23284, Q9BVK5 | Gene names | PPIB, CYPB | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPWD1_MOUSE
|
||||||
| NC score | 0.869499 (rank : 17) | θ value | 7.04741e-37 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CEC6 | Gene names | Ppwd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
PPWD1_HUMAN
|
||||||
| NC score | 0.868998 (rank : 18) | θ value | 1.08726e-37 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96BP3, Q15002, Q7KZ89 | Gene names | PPWD1, KIAA0073 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.811108 (rank : 19) | θ value | 2.12685e-25 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.806321 (rank : 20) | θ value | 1.99067e-23 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.804343 (rank : 21) | θ value | 8.08199e-25 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.768926 (rank : 22) | θ value | 1.99067e-23 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPIE_MOUSE
|
||||||
| NC score | 0.762631 (rank : 23) | θ value | 2.51237e-26 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIE_HUMAN
|
||||||
| NC score | 0.762372 (rank : 24) | θ value | 1.92365e-26 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.750702 (rank : 25) | θ value | 7.56453e-23 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
PPIL4_MOUSE
|
||||||
| NC score | 0.726361 (rank : 26) | θ value | 5.23862e-24 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
PPIL4_HUMAN
|
||||||
| NC score | 0.722707 (rank : 27) | θ value | 4.01107e-24 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.667723 (rank : 28) | θ value | 1.37858e-24 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.660428 (rank : 29) | θ value | 1.80048e-24 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
RBP26_HUMAN
|
||||||
| NC score | 0.111479 (rank : 30) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
RGPD8_HUMAN
|
||||||
| NC score | 0.111256 (rank : 31) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
RBP22_HUMAN
|
||||||
| NC score | 0.110859 (rank : 32) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RANG_MOUSE
|
||||||
| NC score | 0.108385 (rank : 33) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34022, P34023, Q3TL81, Q9D894, Q9DCA3 | Gene names | Ranbp1, Htf9-a, Htf9a | |||
|
Domain Architecture |

|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RANBP1) (HpaII tiny fragments locus 9a protein). | |||||
|
RANG_HUMAN
|
||||||
| NC score | 0.107608 (rank : 34) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43487 | Gene names | RANBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RanBP1). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.089088 (rank : 35) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
FUSIP_HUMAN
|
||||||
| NC score | 0.062002 (rank : 36) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| NC score | 0.062002 (rank : 37) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
SRR35_HUMAN
|
||||||
| NC score | 0.056964 (rank : 38) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |

|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
TRA2A_HUMAN
|
||||||
| NC score | 0.054547 (rank : 39) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |

|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
TRA2A_MOUSE
|
||||||
| NC score | 0.052680 (rank : 40) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PFR5 | Gene names | Tra2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
SFRS2_HUMAN
|
||||||
| NC score | 0.052110 (rank : 41) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| NC score | 0.052110 (rank : 42) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.051913 (rank : 43) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
TRA2B_HUMAN
|
||||||
| NC score | 0.051890 (rank : 44) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| NC score | 0.051890 (rank : 45) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
TF3C1_HUMAN
|
||||||
| NC score | 0.044892 (rank : 46) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.039675 (rank : 47) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
TF3C1_MOUSE
|
||||||
| NC score | 0.035333 (rank : 48) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K284, Q8CAL9 | Gene names | Gtf3c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
MED12_HUMAN
|
||||||
| NC score | 0.012197 (rank : 49) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
K1267_HUMAN
|
||||||
| NC score | 0.009596 (rank : 50) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3B3, Q6AW85, Q8IYH1, Q9NTE7, Q9UFT0, Q9ULF3 | Gene names | KIAA1267 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1267. | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.008527 (rank : 51) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.008343 (rank : 52) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SRPK2_HUMAN
|
||||||
| NC score | 0.007792 (rank : 53) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78362, O75220, O75221, Q6NUL0, Q6V1X2, Q8IYQ3 | Gene names | SRPK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.005917 (rank : 54) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.005293 (rank : 55) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
AN32A_HUMAN
|
||||||
| NC score | 0.004582 (rank : 56) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
MMP16_MOUSE
|
||||||
| NC score | 0.001705 (rank : 57) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
PRD15_HUMAN
|
||||||
| NC score | -0.003662 (rank : 58) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||