Please be patient as the page loads
|
PPIA_MOUSE
|
||||||
| SwissProt Accessions | P17742, Q9CWJ5, Q9R137 | Gene names | Ppia | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein) (SP18). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PPIA_MOUSE
|
||||||
| θ value | 2.38316e-93 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P17742, Q9CWJ5, Q9R137 | Gene names | Ppia | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein) (SP18). | |||||
|
PPIA_HUMAN
|
||||||
| θ value | 3.22093e-90 (rank : 2) | NC score | 0.999904 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P62937, P05092, Q6IBU5, Q96IX3, Q9BRU4, Q9BTY9, Q9UC61 | Gene names | PPIA, CYPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein). | |||||
|
PPIF_MOUSE
|
||||||
| θ value | 1.60597e-73 (rank : 3) | NC score | 0.998540 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KR7 | Gene names | Ppif | |||
|
Domain Architecture |
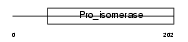
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPIF_HUMAN
|
||||||
| θ value | 3.02872e-72 (rank : 4) | NC score | 0.997985 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30405, Q5W131 | Gene names | PPIF, CYP3 | |||
|
Domain Architecture |
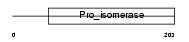
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPIE_HUMAN
|
||||||
| θ value | 4.38363e-63 (rank : 5) | NC score | 0.854135 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |
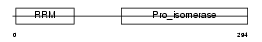
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIE_MOUSE
|
||||||
| θ value | 4.38363e-63 (rank : 6) | NC score | 0.853826 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |
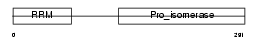
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 1.84173e-61 (rank : 7) | NC score | 0.754530 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 1.84173e-61 (rank : 8) | NC score | 0.762869 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
PPIB_HUMAN
|
||||||
| θ value | 4.85792e-54 (rank : 9) | NC score | 0.989358 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23284, Q9BVK5 | Gene names | PPIB, CYPB | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIB_MOUSE
|
||||||
| θ value | 9.16162e-53 (rank : 10) | NC score | 0.988860 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24369 | Gene names | Ppib | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPID_MOUSE
|
||||||
| θ value | 1.72781e-51 (rank : 11) | NC score | 0.895651 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
PPID_HUMAN
|
||||||
| θ value | 1.11993e-50 (rank : 12) | NC score | 0.892238 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPIC_MOUSE
|
||||||
| θ value | 1.36902e-48 (rank : 13) | NC score | 0.988445 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30412 | Gene names | Ppic, Cypc | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPIC_HUMAN
|
||||||
| θ value | 2.58187e-47 (rank : 14) | NC score | 0.987653 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P45877 | Gene names | PPIC, CYPC | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPIH_HUMAN
|
||||||
| θ value | 1.28137e-46 (rank : 15) | NC score | 0.989767 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43447 | Gene names | PPIH, CYP20, CYPH | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H) (U-snRNP-associated cyclophilin SnuCyp-20) (USA-CYP) (Small nuclear ribonucleoprotein particle-specific cyclophilin H) (CypH). | |||||
|
PPIH_MOUSE
|
||||||
| θ value | 3.48949e-44 (rank : 16) | NC score | 0.988412 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D868, Q9CQU7 | Gene names | Ppih | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 6.15952e-41 (rank : 17) | NC score | 0.821251 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 18) | NC score | 0.839092 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 2.34062e-40 (rank : 19) | NC score | 0.880923 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPIL1_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 20) | NC score | 0.952626 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y3C6, O15001 | Gene names | PPIL1, CYPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 1 (EC 5.2.1.8) (PPIase) (Rotamase). | |||||
|
PPIL1_MOUSE
|
||||||
| θ value | 1.85889e-29 (rank : 21) | NC score | 0.952574 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D0W5, Q3UA03 | Gene names | Ppil1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 1 (EC 5.2.1.8) (PPIase) (Rotamase). | |||||
|
PPWD1_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 22) | NC score | 0.859889 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96BP3, Q15002, Q7KZ89 | Gene names | PPWD1, KIAA0073 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
PPWD1_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 23) | NC score | 0.861958 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CEC6 | Gene names | Ppwd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
PPIL3_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 24) | NC score | 0.937258 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H2H8, Q86WF9, Q96IA9, Q9BXZ1 | Gene names | PPIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 3 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL3) (Cyclophilin J) (CyPJ). | |||||
|
PPIL3_MOUSE
|
||||||
| θ value | 1.62847e-25 (rank : 25) | NC score | 0.936025 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D6L8, Q812D2, Q9CY23 | Gene names | Ppil3, Cyp10l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 3 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL3) (CYP10L). | |||||
|
PPIL2_HUMAN
|
||||||
| θ value | 1.80048e-24 (rank : 26) | NC score | 0.889340 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13356, Q13357, Q8TAH2, Q9BWR8 | Gene names | PPIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
PPIL2_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 27) | NC score | 0.889872 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D787, Q542A2, Q9CZL1 | Gene names | Ppil2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
PPIL4_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 28) | NC score | 0.694722 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
PPIL4_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 29) | NC score | 0.690515 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.003511 (rank : 54) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
CIR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.052792 (rank : 46) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.066355 (rank : 36) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
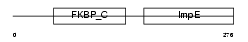
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
FKBP8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.063817 (rank : 40) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
FUSIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.064366 (rank : 38) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.064366 (rank : 39) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
NU153_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.123902 (rank : 35) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
RANG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.149982 (rank : 34) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43487 | Gene names | RANBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RanBP1). | |||||
|
RANG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.151066 (rank : 33) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34022, P34023, Q3TL81, Q9D894, Q9DCA3 | Gene names | Ranbp1, Htf9-a, Htf9a | |||
|
Domain Architecture |
|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RANBP1) (HpaII tiny fragments locus 9a protein). | |||||
|
RBM8A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.050684 (rank : 50) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5S9, Q6FHD1, Q9GZX8, Q9NZI4 | Gene names | RBM8A, RBM8 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A) (RNA-binding protein Y14) (Binder of OVCA1- 1) (BOV-1). | |||||
|
RBM8A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.050684 (rank : 51) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CWZ3, Q9D6U5 | Gene names | Rbm8a, Rbm8 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A). | |||||
|
RBMX2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.054140 (rank : 45) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |
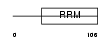
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBP22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.154669 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RBP26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.155519 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.050299 (rank : 53) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
RGPD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.155199 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
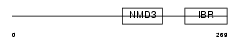
RNF31_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.050371 (rank : 52) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
SFRS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.056029 (rank : 42) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |
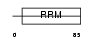
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.056029 (rank : 43) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |
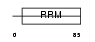
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
SRR35_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.058854 (rank : 41) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |
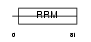
|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
TRA2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.051939 (rank : 47) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |
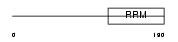
|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
TRA2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.051721 (rank : 48) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.051721 (rank : 49) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
ZRAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.055975 (rank : 44) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
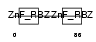
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
ZRAN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.066306 (rank : 37) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
PPIA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.38316e-93 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P17742, Q9CWJ5, Q9R137 | Gene names | Ppia | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein) (SP18). | |||||
|
PPIA_HUMAN
|
||||||
| NC score | 0.999904 (rank : 2) | θ value | 3.22093e-90 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P62937, P05092, Q6IBU5, Q96IX3, Q9BRU4, Q9BTY9, Q9UC61 | Gene names | PPIA, CYPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein). | |||||
|
PPIF_MOUSE
|
||||||
| NC score | 0.998540 (rank : 3) | θ value | 1.60597e-73 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KR7 | Gene names | Ppif | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPIF_HUMAN
|
||||||
| NC score | 0.997985 (rank : 4) | θ value | 3.02872e-72 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30405, Q5W131 | Gene names | PPIF, CYP3 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPIH_HUMAN
|
||||||
| NC score | 0.989767 (rank : 5) | θ value | 1.28137e-46 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43447 | Gene names | PPIH, CYP20, CYPH | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H) (U-snRNP-associated cyclophilin SnuCyp-20) (USA-CYP) (Small nuclear ribonucleoprotein particle-specific cyclophilin H) (CypH). | |||||
|
PPIB_HUMAN
|
||||||
| NC score | 0.989358 (rank : 6) | θ value | 4.85792e-54 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23284, Q9BVK5 | Gene names | PPIB, CYPB | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIB_MOUSE
|
||||||
| NC score | 0.988860 (rank : 7) | θ value | 9.16162e-53 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24369 | Gene names | Ppib | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIC_MOUSE
|
||||||
| NC score | 0.988445 (rank : 8) | θ value | 1.36902e-48 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30412 | Gene names | Ppic, Cypc | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPIH_MOUSE
|
||||||
| NC score | 0.988412 (rank : 9) | θ value | 3.48949e-44 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D868, Q9CQU7 | Gene names | Ppih | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H). | |||||
|
PPIC_HUMAN
|
||||||
| NC score | 0.987653 (rank : 10) | θ value | 2.58187e-47 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P45877 | Gene names | PPIC, CYPC | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPIL1_HUMAN
|
||||||
| NC score | 0.952626 (rank : 11) | θ value | 1.85889e-29 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y3C6, O15001 | Gene names | PPIL1, CYPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 1 (EC 5.2.1.8) (PPIase) (Rotamase). | |||||
|
PPIL1_MOUSE
|
||||||
| NC score | 0.952574 (rank : 12) | θ value | 1.85889e-29 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D0W5, Q3UA03 | Gene names | Ppil1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 1 (EC 5.2.1.8) (PPIase) (Rotamase). | |||||
|
PPIL3_HUMAN
|
||||||
| NC score | 0.937258 (rank : 13) | θ value | 2.51237e-26 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H2H8, Q86WF9, Q96IA9, Q9BXZ1 | Gene names | PPIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 3 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL3) (Cyclophilin J) (CyPJ). | |||||
|
PPIL3_MOUSE
|
||||||
| NC score | 0.936025 (rank : 14) | θ value | 1.62847e-25 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D6L8, Q812D2, Q9CY23 | Gene names | Ppil3, Cyp10l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 3 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL3) (CYP10L). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.895651 (rank : 15) | θ value | 1.72781e-51 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.892238 (rank : 16) | θ value | 1.11993e-50 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPIL2_MOUSE
|
||||||
| NC score | 0.889872 (rank : 17) | θ value | 3.07116e-24 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D787, Q542A2, Q9CZL1 | Gene names | Ppil2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
PPIL2_HUMAN
|
||||||
| NC score | 0.889340 (rank : 18) | θ value | 1.80048e-24 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13356, Q13357, Q8TAH2, Q9BWR8 | Gene names | PPIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 2 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-60) (Cyclophilin-like protein Cyp-60). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.880923 (rank : 19) | θ value | 2.34062e-40 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPWD1_MOUSE
|
||||||
| NC score | 0.861958 (rank : 20) | θ value | 3.87602e-27 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CEC6 | Gene names | Ppwd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
PPWD1_HUMAN
|
||||||
| NC score | 0.859889 (rank : 21) | θ value | 3.87602e-27 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96BP3, Q15002, Q7KZ89 | Gene names | PPWD1, KIAA0073 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
PPIE_HUMAN
|
||||||
| NC score | 0.854135 (rank : 22) | θ value | 4.38363e-63 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIE_MOUSE
|
||||||
| NC score | 0.853826 (rank : 23) | θ value | 4.38363e-63 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.839092 (rank : 24) | θ value | 2.34062e-40 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.821251 (rank : 25) | θ value | 6.15952e-41 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.762869 (rank : 26) | θ value | 1.84173e-61 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.754530 (rank : 27) | θ value | 1.84173e-61 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
PPIL4_MOUSE
|
||||||
| NC score | 0.694722 (rank : 28) | θ value | 7.09661e-13 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
PPIL4_HUMAN
|
||||||
| NC score | 0.690515 (rank : 29) | θ value | 9.26847e-13 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
RBP26_HUMAN
|
||||||
| NC score | 0.155519 (rank : 30) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
RGPD8_HUMAN
|
||||||
| NC score | 0.155199 (rank : 31) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
RBP22_HUMAN
|
||||||
| NC score | 0.154669 (rank : 32) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RANG_MOUSE
|
||||||
| NC score | 0.151066 (rank : 33) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34022, P34023, Q3TL81, Q9D894, Q9DCA3 | Gene names | Ranbp1, Htf9-a, Htf9a | |||
|
Domain Architecture |

|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RANBP1) (HpaII tiny fragments locus 9a protein). | |||||
|
RANG_HUMAN
|
||||||
| NC score | 0.149982 (rank : 34) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43487 | Gene names | RANBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RanBP1). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.123902 (rank : 35) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.066355 (rank : 36) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
ZRAN1_HUMAN
|
||||||
| NC score | 0.066306 (rank : 37) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
FUSIP_HUMAN
|
||||||
| NC score | 0.064366 (rank : 38) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| NC score | 0.064366 (rank : 39) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
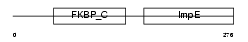
FKBP8_MOUSE
|
||||||
| NC score | 0.063817 (rank : 40) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
SRR35_HUMAN
|
||||||
| NC score | 0.058854 (rank : 41) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |

|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
SFRS2_HUMAN
|
||||||
| NC score | 0.056029 (rank : 42) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| NC score | 0.056029 (rank : 43) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
ZRAB2_HUMAN
|
||||||
| NC score | 0.055975 (rank : 44) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
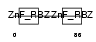
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
RBMX2_MOUSE
|
||||||
| NC score | 0.054140 (rank : 45) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.052792 (rank : 46) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
TRA2A_HUMAN
|
||||||
| NC score | 0.051939 (rank : 47) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |

|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
TRA2B_HUMAN
|
||||||
| NC score | 0.051721 (rank : 48) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| NC score | 0.051721 (rank : 49) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
RBM8A_HUMAN
|
||||||
| NC score | 0.050684 (rank : 50) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5S9, Q6FHD1, Q9GZX8, Q9NZI4 | Gene names | RBM8A, RBM8 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A) (RNA-binding protein Y14) (Binder of OVCA1- 1) (BOV-1). | |||||
|
RBM8A_MOUSE
|
||||||
| NC score | 0.050684 (rank : 51) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CWZ3, Q9D6U5 | Gene names | Rbm8a, Rbm8 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A). | |||||
|
RNF31_HUMAN
|
||||||
| NC score | 0.050371 (rank : 52) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.050299 (rank : 53) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.003511 (rank : 54) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||