Please be patient as the page loads
|
NRDC_MOUSE
|
||||||
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NRDC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.986043 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_MOUSE
|
||||||
| θ value | 2.26584e-152 (rank : 3) | NC score | 0.911024 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| θ value | 3.27187e-151 (rank : 4) | NC score | 0.910588 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 5) | NC score | 0.139209 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 6) | NC score | 0.132573 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MPPA_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 7) | NC score | 0.267838 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |
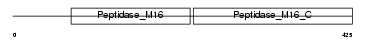
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPB_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 8) | NC score | 0.254847 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |
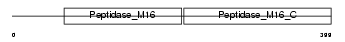
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
UQCR1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 9) | NC score | 0.239931 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |
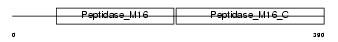
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 10) | NC score | 0.112502 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 11) | NC score | 0.094756 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
UQCR1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 12) | NC score | 0.254158 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |
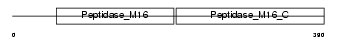
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPB_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 13) | NC score | 0.239505 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.236334 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 15) | NC score | 0.100855 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 16) | NC score | 0.014500 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.108053 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
GPTC3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.066325 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.085363 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PREP_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.126543 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K411, Q3THC4, Q3TJI1, Q3TPP0, Q3URQ8, Q4KUG2, Q4KUG3, Q6ZPX6, Q922N1, Q9CV63 | Gene names | Pitrm1, Kiaa1104, Ntup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (Pitrilysin metalloproteinase 1). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.058264 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.037794 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
PREP_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.132666 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5JRX3, O95204, Q2M2G6, Q4VBR1, Q5JRW7, Q7L5Z7, Q9BSI6, Q9BVJ5, Q9UPP8 | Gene names | PITRM1, KIAA1104, MP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (hPreP) (Pitrilysin metalloproteinase 1) (Metalloprotease 1) (hMP1). | |||||
|
RN139_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.037032 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WU17, O75485, Q7LDL3 | Gene names | RNF139, TRC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 139 (Translocation in renal carcinoma on chromosome 8). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.027860 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.067990 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
CALX_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.058882 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CALX_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.057981 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.091165 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.081572 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.011785 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
TINAL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.051642 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.051823 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
F10A4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.042753 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.039748 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.010791 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
F10A5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.043131 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
CLGN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.043951 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor. | |||||
|
F10A1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.041478 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
RAG2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.051237 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.010753 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.010926 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
TSYL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.020900 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H489, Q3MJ63, Q3MJ65 | Gene names | TSPYL3 | |||
|
Domain Architecture |
|
|||||
| Description | Testis-specific Y-encoded-like protein 3 (TSPY-like 3). | |||||
|
LYST_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.013911 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
RAG2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.039715 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55895, Q8TBL4 | Gene names | RAG2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
SV2C_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.015703 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q496J9, Q496K1, Q9UPU8 | Gene names | SV2C, KIAA1054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle glycoprotein 2C. | |||||
|
TINAL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.043365 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
ASPH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.048284 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.022896 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
RBBP5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.017907 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.020413 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
EZH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.015735 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
HDGR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.034084 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.044276 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.023667 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
SEPT8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.012777 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92599, Q8IX36, Q8IX37, Q9BVB3 | Gene names | SEPT8, KIAA0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
STAG2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.021465 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N3U4, O00540, Q5JTI6, Q9H1N8 | Gene names | STAG2, SA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
STAG2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.021508 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35638, Q6NZN7 | Gene names | Stag2, Sa2, Sap2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.016033 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
ULE1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.018813 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.045720 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.044026 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.005504 (rank : 111) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
SG16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.056449 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
41_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.009574 (rank : 102) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
CCD82_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.052210 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.069289 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.059275 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.019147 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.008026 (rank : 104) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.025245 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.006660 (rank : 108) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DCDC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.027870 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.064116 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
EZH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.013561 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
IF2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.018760 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20042, Q9BVU0, Q9UJE4 | Gene names | EIF2S2, EIF2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 2 (Eukaryotic translation initiation factor 2 subunit beta) (eIF-2-beta). | |||||
|
ILF3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.010586 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.042730 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
SUCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.027977 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUM5, Q8C2C3, Q9DBA3, Q9DCI8 | Gene names | Suclg1 | |||
|
Domain Architecture |
|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.009280 (rank : 103) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
TTLL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.013807 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |
|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.039824 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.007100 (rank : 107) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
GRP78_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.005969 (rank : 110) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.006022 (rank : 109) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
IGHG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.003007 (rank : 113) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01857 | Gene names | IGHG1 | |||
|
Domain Architecture |
|
|||||
| Description | Ig gamma-1 chain C region. | |||||
|
LYRIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.020950 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.023213 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.009844 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.016457 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.017125 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PO210_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.007304 (rank : 106) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEM1, O94980, Q6NXG6, Q8NBJ1, Q9H6C8, Q9UFP3 | Gene names | NUP210, KIAA0906 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore membrane glycoprotein 210 precursor (POM210) (Nuclear pore protein gp210). | |||||
|
PWP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.004869 (rank : 112) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LL5, Q9D6T6 | Gene names | Pwp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periodic tryptophan protein 1 homolog. | |||||
|
SEPT8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.007889 (rank : 105) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHH9, Q80YC7, Q9ESF7 | Gene names | Sept8, Kiaa0202 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-8. | |||||
|
SUCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.024917 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53597, Q9BWB0, Q9UNP6 | Gene names | SUCLG1 | |||
|
Domain Architecture |
|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
ZSCA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | -0.001256 (rank : 114) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.052364 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
AN32B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.052625 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.052635 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.051862 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.058061 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058363 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.071547 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.067871 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.071137 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.111867 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.109861 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.074987 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.089074 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
SRCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.057595 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.051349 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
UQCR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.086107 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.087604 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055062 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_HUMAN
|
||||||
| NC score | 0.986043 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_MOUSE
|
||||||
| NC score | 0.911024 (rank : 3) | θ value | 2.26584e-152 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| NC score | 0.910588 (rank : 4) | θ value | 3.27187e-151 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
MPPA_MOUSE
|
||||||
| NC score | 0.267838 (rank : 5) | θ value | 4.1701e-05 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPB_HUMAN
|
||||||
| NC score | 0.254847 (rank : 6) | θ value | 0.000270298 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
UQCR1_HUMAN
|
||||||
| NC score | 0.254158 (rank : 7) | θ value | 0.000461057 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_MOUSE
|
||||||
| NC score | 0.239931 (rank : 8) | θ value | 0.000270298 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPB_MOUSE
|
||||||
| NC score | 0.239505 (rank : 9) | θ value | 0.000786445 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_HUMAN
|
||||||
| NC score | 0.236334 (rank : 10) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.139209 (rank : 11) | θ value | 4.45536e-07 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PREP_HUMAN
|
||||||
| NC score | 0.132666 (rank : 12) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5JRX3, O95204, Q2M2G6, Q4VBR1, Q5JRW7, Q7L5Z7, Q9BSI6, Q9BVJ5, Q9UPP8 | Gene names | PITRM1, KIAA1104, MP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (hPreP) (Pitrilysin metalloproteinase 1) (Metalloprotease 1) (hMP1). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.132573 (rank : 13) | θ value | 1.09739e-05 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PREP_MOUSE
|
||||||
| NC score | 0.126543 (rank : 14) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K411, Q3THC4, Q3TJI1, Q3TPP0, Q3URQ8, Q4KUG2, Q4KUG3, Q6ZPX6, Q922N1, Q9CV63 | Gene names | Pitrm1, Kiaa1104, Ntup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (Pitrilysin metalloproteinase 1). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.112502 (rank : 15) | θ value | 0.00035302 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.111867 (rank : 16) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.109861 (rank : 17) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.108053 (rank : 18) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.100855 (rank : 19) | θ value | 0.0113563 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.094756 (rank : 20) | θ value | 0.000461057 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.091165 (rank : 21) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.089074 (rank : 22) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
UQCR2_MOUSE
|
||||||
| NC score | 0.087604 (rank : 23) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_HUMAN
|
||||||
| NC score | 0.086107 (rank : 24) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.085363 (rank : 25) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.081572 (rank : 26) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.074987 (rank : 27) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.071547 (rank : 28) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.071137 (rank : 29) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.069289 (rank : 30) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.067990 (rank : 31) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.067871 (rank : 32) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
GPTC3_HUMAN
|
||||||
| NC score | 0.066325 (rank : 33) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.064116 (rank : 34) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.059275 (rank : 35) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.058882 (rank : 36) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.058363 (rank : 37) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.058264 (rank : 38) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.058061 (rank : 39) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CALX_MOUSE
|
||||||
| NC score | 0.057981 (rank : 40) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.057595 (rank : 41) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
SG16_MOUSE
|
||||||
| NC score | 0.056449 (rank : 42) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.055062 (rank : 43) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.052635 (rank : 44) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.052625 (rank : 45) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.052364 (rank : 46) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
CCD82_HUMAN
|
||||||
| NC score | 0.052210 (rank : 47) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.051862 (rank : 48) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.051823 (rank : 49) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
TINAL_MOUSE
|
||||||
| NC score | 0.051642 (rank : 50) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.051349 (rank : 51) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
RAG2_MOUSE
|
||||||
| NC score | 0.051237 (rank : 52) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
ASPH_HUMAN
|
||||||
| NC score | 0.048284 (rank : 53) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.045720 (rank : 54) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.044276 (rank : 55) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.044026 (rank : 56) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
CLGN_HUMAN
|
||||||
| NC score | 0.043951 (rank : 57) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor. | |||||
|
TINAL_HUMAN
|
||||||
| NC score | 0.043365 (rank : 58) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.043131 (rank : 59) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.042753 (rank : 60) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.042730 (rank : 61) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.041478 (rank : 62) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.039824 (rank : 63) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.039748 (rank : 64) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
RAG2_HUMAN
|
||||||
| NC score | 0.039715 (rank : 65) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55895, Q8TBL4 | Gene names | RAG2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.037794 (rank : 66) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
RN139_HUMAN
|
||||||
| NC score | 0.037032 (rank : 67) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WU17, O75485, Q7LDL3 | Gene names | RNF139, TRC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 139 (Translocation in renal carcinoma on chromosome 8). | |||||
|
HDGR3_HUMAN
|
||||||
| NC score | 0.034084 (rank : 68) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
SUCA_MOUSE
|
||||||
| NC score | 0.027977 (rank : 69) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUM5, Q8C2C3, Q9DBA3, Q9DCI8 | Gene names | Suclg1 | |||
|
Domain Architecture |

|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
DCDC2_MOUSE
|
||||||
| NC score | 0.027870 (rank : 70) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.027860 (rank : 71) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.025245 (rank : 72) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SUCA_HUMAN
|
||||||
| NC score | 0.024917 (rank : 73) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53597, Q9BWB0, Q9UNP6 | Gene names | SUCLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.023667 (rank : 74) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.023213 (rank : 75) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.022896 (rank : 76) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
STAG2_MOUSE
|
||||||
| NC score | 0.021508 (rank : 77) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35638, Q6NZN7 | Gene names | Stag2, Sa2, Sap2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
STAG2_HUMAN
|
||||||
| NC score | 0.021465 (rank : 78) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N3U4, O00540, Q5JTI6, Q9H1N8 | Gene names | STAG2, SA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
LYRIC_HUMAN
|
||||||
| NC score | 0.020950 (rank : 79) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
TSYL3_HUMAN
|
||||||
| NC score | 0.020900 (rank : 80) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H489, Q3MJ63, Q3MJ65 | Gene names | TSPYL3 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific Y-encoded-like protein 3 (TSPY-like 3). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.020413 (rank : 81) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.019147 (rank : 82) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ULE1B_MOUSE
|
||||||
| NC score | 0.018813 (rank : 83) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
IF2B_HUMAN
|
||||||
| NC score | 0.018760 (rank : 84) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20042, Q9BVU0, Q9UJE4 | Gene names | EIF2S2, EIF2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 2 (Eukaryotic translation initiation factor 2 subunit beta) (eIF-2-beta). | |||||
|
RBBP5_HUMAN
|
||||||
| NC score | 0.017907 (rank : 85) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.017125 (rank : 86) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.016457 (rank : 87) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.016033 (rank : 88) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
EZH1_HUMAN
|
||||||
| NC score | 0.015735 (rank : 89) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
SV2C_HUMAN
|
||||||
| NC score | 0.015703 (rank : 90) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q496J9, Q496K1, Q9UPU8 | Gene names | SV2C, KIAA1054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle glycoprotein 2C. | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.014500 (rank : 91) | θ value | 0.0148317 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.013911 (rank : 92) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
TTLL1_HUMAN
|
||||||
| NC score | 0.013807 (rank : 93) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |

|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
EZH1_MOUSE
|
||||||
| NC score | 0.013561 (rank : 94) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
SEPT8_HUMAN
|
||||||
| NC score | 0.012777 (rank : 95) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92599, Q8IX36, Q8IX37, Q9BVB3 | Gene names | SEPT8, KIAA0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.011785 (rank : 96) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.010926 (rank : 97) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.010791 (rank : 98) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.010753 (rank : 99) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
ILF3_MOUSE
|
||||||
| NC score | 0.010586 (rank : 100) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.009844 (rank : 101) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.009574 (rank : 102) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.009280 (rank : 103) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.008026 (rank : 104) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
SEPT8_MOUSE
|
||||||
| NC score | 0.007889 (rank : 105) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHH9, Q80YC7, Q9ESF7 | Gene names | Sept8, Kiaa0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
PO210_HUMAN
|
||||||
| NC score | 0.007304 (rank : 106) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEM1, O94980, Q6NXG6, Q8NBJ1, Q9H6C8, Q9UFP3 | Gene names | NUP210, KIAA0906 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore membrane glycoprotein 210 precursor (POM210) (Nuclear pore protein gp210). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.007100 (rank : 107) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.006660 (rank : 108) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GRP78_MOUSE
|
||||||
| NC score | 0.006022 (rank : 109) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
GRP78_HUMAN
|
||||||
| NC score | 0.005969 (rank : 110) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.005504 (rank : 111) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
PWP1_MOUSE
|
||||||
| NC score | 0.004869 (rank : 112) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LL5, Q9D6T6 | Gene names | Pwp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periodic tryptophan protein 1 homolog. | |||||
|
IGHG1_HUMAN
|
||||||
| NC score | 0.003007 (rank : 113) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01857 | Gene names | IGHG1 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-1 chain C region. | |||||
|
ZSCA5_HUMAN
|
||||||
| NC score | -0.001256 (rank : 114) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||