Please be patient as the page loads
|
ULE1B_MOUSE
|
||||||
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ULE1B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990613 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULE1B_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBA3_HUMAN
|
||||||
| θ value | 4.41421e-39 (rank : 3) | NC score | 0.826839 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_MOUSE
|
||||||
| θ value | 5.76516e-39 (rank : 4) | NC score | 0.826943 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBE1_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 5) | NC score | 0.722730 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UBE1_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 6) | NC score | 0.721015 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
UBE1L_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 7) | NC score | 0.714179 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
MOCS3_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 8) | NC score | 0.669466 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |
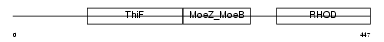
|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
UE1D1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 9) | NC score | 0.494484 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
UE1D1_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 10) | NC score | 0.485163 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
ULE1A_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 11) | NC score | 0.561713 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |
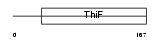
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 12) | NC score | 0.572730 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |
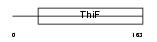
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULA1_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 13) | NC score | 0.462197 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
ULA1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 14) | NC score | 0.456089 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ATG7_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 15) | NC score | 0.237896 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
ATG7_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 16) | NC score | 0.234667 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
NEK1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.010982 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |
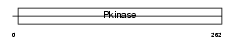
|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.015897 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.022048 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.041176 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DAXX_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.042335 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.038097 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.024102 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
API5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.049871 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZZ5, O15441, Q9Y4J7 | Gene names | API5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis inhibitor 5 (API-5) (Fibroblast growth factor 2-interacting factor) (FIF) (Protein XAGL) (Antiapoptosis clone 11 protein) (AAC- 11). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.023195 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.009820 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.028887 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
FOXD3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.006489 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61060 | Gene names | Foxd3, Hfh2 | |||
|
Domain Architecture |
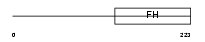
|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis) (Hepatocyte nuclear factor 3 forkhead homolog 2) (HFH-2). | |||||
|
NOL7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.030219 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7Z3, Q9CY79, Q9JJE2 | Gene names | Nol7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 7. | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.018813 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.021068 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.032020 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.024456 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
GASP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.017066 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
IFI4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.013074 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15092 | Gene names | Ifi204 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 204 (Ifi-204) (Interferon-inducible protein p204). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.019768 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
XRCC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.030820 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.029645 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
CU025_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.018954 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y426, Q5R2V7, Q6AHX8, Q9NSE6 | Gene names | C21orf25, C21orf258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf25 precursor. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.029978 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.018557 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.015545 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
NUP37_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.017149 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWU9 | Gene names | Nup37 | |||
|
Domain Architecture |
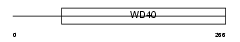
|
|||||
| Description | Nucleoporin Nup37. | |||||
|
PESC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.017963 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00541, Q6IC29 | Gene names | PES1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
API5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.035988 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35841, Q922L2 | Gene names | Api5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis inhibitor 5 (API-5) (AAC-11). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.014194 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
PTMA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.030912 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
CALU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.010713 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.008599 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.008531 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.025285 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NUP37_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.015435 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFH4, Q9H644 | Gene names | NUP37 | |||
|
Domain Architecture |
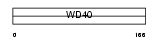
|
|||||
| Description | Nucleoporin Nup37 (p37). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.006055 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.016674 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TAF11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.014890 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JX1, Q3UZH1, Q9CT98 | Gene names | Taf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 11 (Transcription initiation factor TFIID 28 kDa subunit) (TAF(II)28) (TAFII-28) (TAFII28) (TFIID subunit p30-beta). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.016466 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ULE1B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULE1B_HUMAN
|
||||||
| NC score | 0.990613 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBA3_MOUSE
|
||||||
| NC score | 0.826943 (rank : 3) | θ value | 5.76516e-39 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_HUMAN
|
||||||
| NC score | 0.826839 (rank : 4) | θ value | 4.41421e-39 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBE1_HUMAN
|
||||||
| NC score | 0.722730 (rank : 5) | θ value | 2.86786e-30 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UBE1_MOUSE
|
||||||
| NC score | 0.721015 (rank : 6) | θ value | 4.89182e-30 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
UBE1L_HUMAN
|
||||||
| NC score | 0.714179 (rank : 7) | θ value | 1.99067e-23 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
MOCS3_HUMAN
|
||||||
| NC score | 0.669466 (rank : 8) | θ value | 2.88119e-14 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |

|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
ULE1A_HUMAN
|
||||||
| NC score | 0.572730 (rank : 9) | θ value | 4.76016e-09 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_MOUSE
|
||||||
| NC score | 0.561713 (rank : 10) | θ value | 2.79066e-09 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
UE1D1_HUMAN
|
||||||
| NC score | 0.494484 (rank : 11) | θ value | 2.28291e-11 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
UE1D1_MOUSE
|
||||||
| NC score | 0.485163 (rank : 12) | θ value | 1.9326e-10 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
ULA1_MOUSE
|
||||||
| NC score | 0.462197 (rank : 13) | θ value | 1.99992e-07 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
ULA1_HUMAN
|
||||||
| NC score | 0.456089 (rank : 14) | θ value | 7.59969e-07 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ATG7_MOUSE
|
||||||
| NC score | 0.237896 (rank : 15) | θ value | 0.000121331 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
ATG7_HUMAN
|
||||||
| NC score | 0.234667 (rank : 16) | θ value | 0.00020696 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
API5_HUMAN
|
||||||
| NC score | 0.049871 (rank : 17) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZZ5, O15441, Q9Y4J7 | Gene names | API5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis inhibitor 5 (API-5) (Fibroblast growth factor 2-interacting factor) (FIF) (Protein XAGL) (Antiapoptosis clone 11 protein) (AAC- 11). | |||||
|
DAXX_MOUSE
|
||||||
| NC score | 0.042335 (rank : 18) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.041176 (rank : 19) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.038097 (rank : 20) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
API5_MOUSE
|
||||||
| NC score | 0.035988 (rank : 21) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35841, Q922L2 | Gene names | Api5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis inhibitor 5 (API-5) (AAC-11). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.032020 (rank : 22) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.030912 (rank : 23) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
XRCC1_HUMAN
|
||||||
| NC score | 0.030820 (rank : 24) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
NOL7_MOUSE
|
||||||
| NC score | 0.030219 (rank : 25) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7Z3, Q9CY79, Q9JJE2 | Gene names | Nol7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 7. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.029978 (rank : 26) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.029645 (rank : 27) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.028887 (rank : 28) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.025285 (rank : 29) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.024456 (rank : 30) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.024102 (rank : 31) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.023195 (rank : 32) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.022048 (rank : 33) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.021068 (rank : 34) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.019768 (rank : 35) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
CU025_HUMAN
|
||||||
| NC score | 0.018954 (rank : 36) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y426, Q5R2V7, Q6AHX8, Q9NSE6 | Gene names | C21orf25, C21orf258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf25 precursor. | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.018813 (rank : 37) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.018557 (rank : 38) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
PESC_HUMAN
|
||||||
| NC score | 0.017963 (rank : 39) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00541, Q6IC29 | Gene names | PES1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
NUP37_MOUSE
|
||||||
| NC score | 0.017149 (rank : 40) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWU9 | Gene names | Nup37 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin Nup37. | |||||
|
GASP1_HUMAN
|
||||||
| NC score | 0.017066 (rank : 41) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.016674 (rank : 42) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.016466 (rank : 43) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.015897 (rank : 44) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.015545 (rank : 45) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
NUP37_HUMAN
|
||||||
| NC score | 0.015435 (rank : 46) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFH4, Q9H644 | Gene names | NUP37 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin Nup37 (p37). | |||||
|
TAF11_MOUSE
|
||||||
| NC score | 0.014890 (rank : 47) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JX1, Q3UZH1, Q9CT98 | Gene names | Taf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 11 (Transcription initiation factor TFIID 28 kDa subunit) (TAF(II)28) (TAFII-28) (TAFII28) (TFIID subunit p30-beta). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.014194 (rank : 48) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
IFI4_MOUSE
|
||||||
| NC score | 0.013074 (rank : 49) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15092 | Gene names | Ifi204 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 204 (Ifi-204) (Interferon-inducible protein p204). | |||||
|
NEK1_HUMAN
|
||||||
| NC score | 0.010982 (rank : 50) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
CALU_HUMAN
|
||||||
| NC score | 0.010713 (rank : 51) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.009820 (rank : 52) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.008599 (rank : 53) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.008531 (rank : 54) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
FOXD3_MOUSE
|
||||||
| NC score | 0.006489 (rank : 55) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61060 | Gene names | Foxd3, Hfh2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis) (Hepatocyte nuclear factor 3 forkhead homolog 2) (HFH-2). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.006055 (rank : 56) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||