Please be patient as the page loads
|
ULE1B_HUMAN
|
||||||
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ULE1B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULE1B_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990613 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBA3_HUMAN
|
||||||
| θ value | 2.58786e-39 (rank : 3) | NC score | 0.830515 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 4) | NC score | 0.830444 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBE1_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 5) | NC score | 0.720220 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UBE1_MOUSE
|
||||||
| θ value | 2.68423e-28 (rank : 6) | NC score | 0.718093 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
UBE1L_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 7) | NC score | 0.706471 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
MOCS3_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 8) | NC score | 0.643794 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |
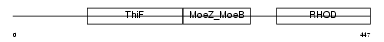
|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
ULE1A_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 9) | NC score | 0.559723 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |
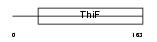
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 10) | NC score | 0.549650 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |
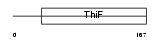
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULA1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 11) | NC score | 0.457744 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
ULA1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 12) | NC score | 0.451428 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
UE1D1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 13) | NC score | 0.437992 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
UE1D1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 14) | NC score | 0.428095 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
ATG7_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.207379 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
ATG7_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.203852 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.010954 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
API5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.051502 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZZ5, O15441, Q9Y4J7 | Gene names | API5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis inhibitor 5 (API-5) (Fibroblast growth factor 2-interacting factor) (FIF) (Protein XAGL) (Antiapoptosis clone 11 protein) (AAC- 11). | |||||
|
BSCL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.021177 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.007798 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.004082 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
K0586_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.016589 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.004441 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
API5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.037235 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35841, Q922L2 | Gene names | Api5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis inhibitor 5 (API-5) (AAC-11). | |||||
|
GASP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.015511 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.003147 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.007559 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
CALU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.010761 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
RRP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.011496 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.006519 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.001874 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
ULE1B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULE1B_MOUSE
|
||||||
| NC score | 0.990613 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBA3_HUMAN
|
||||||
| NC score | 0.830515 (rank : 3) | θ value | 2.58786e-39 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_MOUSE
|
||||||
| NC score | 0.830444 (rank : 4) | θ value | 3.37985e-39 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBE1_HUMAN
|
||||||
| NC score | 0.720220 (rank : 5) | θ value | 2.42779e-29 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UBE1_MOUSE
|
||||||
| NC score | 0.718093 (rank : 6) | θ value | 2.68423e-28 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
UBE1L_HUMAN
|
||||||
| NC score | 0.706471 (rank : 7) | θ value | 3.51386e-20 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
MOCS3_HUMAN
|
||||||
| NC score | 0.643794 (rank : 8) | θ value | 1.9326e-10 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |

|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
ULE1A_HUMAN
|
||||||
| NC score | 0.559723 (rank : 9) | θ value | 3.08544e-08 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_MOUSE
|
||||||
| NC score | 0.549650 (rank : 10) | θ value | 5.81887e-07 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULA1_MOUSE
|
||||||
| NC score | 0.457744 (rank : 11) | θ value | 9.92553e-07 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
ULA1_HUMAN
|
||||||
| NC score | 0.451428 (rank : 12) | θ value | 3.77169e-06 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
UE1D1_HUMAN
|
||||||
| NC score | 0.437992 (rank : 13) | θ value | 8.40245e-06 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
UE1D1_MOUSE
|
||||||
| NC score | 0.428095 (rank : 14) | θ value | 7.1131e-05 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
ATG7_MOUSE
|
||||||
| NC score | 0.207379 (rank : 15) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
ATG7_HUMAN
|
||||||
| NC score | 0.203852 (rank : 16) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
API5_HUMAN
|
||||||
| NC score | 0.051502 (rank : 17) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZZ5, O15441, Q9Y4J7 | Gene names | API5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis inhibitor 5 (API-5) (Fibroblast growth factor 2-interacting factor) (FIF) (Protein XAGL) (Antiapoptosis clone 11 protein) (AAC- 11). | |||||
|
API5_MOUSE
|
||||||
| NC score | 0.037235 (rank : 18) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35841, Q922L2 | Gene names | Api5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis inhibitor 5 (API-5) (AAC-11). | |||||
|
BSCL2_MOUSE
|
||||||
| NC score | 0.021177 (rank : 19) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.016589 (rank : 20) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
GASP1_HUMAN
|
||||||
| NC score | 0.015511 (rank : 21) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.011496 (rank : 22) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.010954 (rank : 23) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
CALU_HUMAN
|
||||||
| NC score | 0.010761 (rank : 24) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.007798 (rank : 25) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.007559 (rank : 26) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.006519 (rank : 27) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.004441 (rank : 28) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.004082 (rank : 29) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.003147 (rank : 30) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.001874 (rank : 31) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||