Please be patient as the page loads
|
ULA1_MOUSE
|
||||||
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ULA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999091 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ULA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
ULE1A_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 3) | NC score | 0.712726 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |
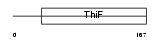
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 4) | NC score | 0.705935 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |
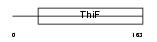
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
UBE1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 5) | NC score | 0.560876 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UBE1_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 6) | NC score | 0.556430 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
UBE1L_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 7) | NC score | 0.562917 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
ULE1B_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 8) | NC score | 0.462197 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULE1B_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 9) | NC score | 0.457744 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBA3_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 10) | NC score | 0.459511 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 11) | NC score | 0.459233 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
MOCS3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 12) | NC score | 0.366413 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |
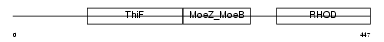
|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
CJ086_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.068365 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
PKHG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.026838 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
UE1D1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.176413 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.006953 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.000954 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
AG10A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.017139 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5BKT4, Q6NS98, Q96DU0, Q96SM6 | Gene names | ALG10, ALG10A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,2-glucosyltransferase ALG10-A (EC 2.4.1.-) (Alpha-2- glucosyltransferase ALG10-A) (Asparagine-linked glycosylation protein 10 homolog A). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.006895 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.000699 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.007508 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
SYCP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.021732 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
ATG7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.051013 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
UE1D1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.151396 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
ULA1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
ULA1_HUMAN
|
||||||
| NC score | 0.999091 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ULE1A_MOUSE
|
||||||
| NC score | 0.712726 (rank : 3) | θ value | 5.42112e-21 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_HUMAN
|
||||||
| NC score | 0.705935 (rank : 4) | θ value | 2.69047e-20 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
UBE1L_HUMAN
|
||||||
| NC score | 0.562917 (rank : 5) | θ value | 1.02475e-11 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
UBE1_HUMAN
|
||||||
| NC score | 0.560876 (rank : 6) | θ value | 7.09661e-13 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UBE1_MOUSE
|
||||||
| NC score | 0.556430 (rank : 7) | θ value | 1.2105e-12 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
ULE1B_MOUSE
|
||||||
| NC score | 0.462197 (rank : 8) | θ value | 1.99992e-07 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBA3_HUMAN
|
||||||
| NC score | 0.459511 (rank : 9) | θ value | 1.69304e-06 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_MOUSE
|
||||||
| NC score | 0.459233 (rank : 10) | θ value | 1.69304e-06 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
ULE1B_HUMAN
|
||||||
| NC score | 0.457744 (rank : 11) | θ value | 9.92553e-07 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
MOCS3_HUMAN
|
||||||
| NC score | 0.366413 (rank : 12) | θ value | 0.0193708 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |

|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
UE1D1_HUMAN
|
||||||
| NC score | 0.176413 (rank : 13) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
UE1D1_MOUSE
|
||||||
| NC score | 0.151396 (rank : 14) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
CJ086_HUMAN
|
||||||
| NC score | 0.068365 (rank : 15) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
ATG7_HUMAN
|
||||||
| NC score | 0.051013 (rank : 16) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
PKHG1_HUMAN
|
||||||
| NC score | 0.026838 (rank : 17) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
SYCP2_HUMAN
|
||||||
| NC score | 0.021732 (rank : 18) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
AG10A_HUMAN
|
||||||
| NC score | 0.017139 (rank : 19) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5BKT4, Q6NS98, Q96DU0, Q96SM6 | Gene names | ALG10, ALG10A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,2-glucosyltransferase ALG10-A (EC 2.4.1.-) (Alpha-2- glucosyltransferase ALG10-A) (Asparagine-linked glycosylation protein 10 homolog A). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.007508 (rank : 20) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.006953 (rank : 21) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.006895 (rank : 22) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.000954 (rank : 23) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.000699 (rank : 24) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||