Please be patient as the page loads
|
MOCS3_HUMAN
|
||||||
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |
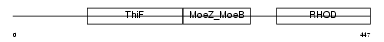
|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MOCS3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |

|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
ULE1B_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 2) | NC score | 0.669466 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBE1_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 3) | NC score | 0.609982 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
ULE1B_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 4) | NC score | 0.643794 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBE1L_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 5) | NC score | 0.617097 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
UBE1_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 6) | NC score | 0.604913 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UBA3_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 7) | NC score | 0.610921 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 8) | NC score | 0.609670 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UE1D1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 9) | NC score | 0.503647 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
UE1D1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 10) | NC score | 0.502997 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
ULE1A_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 11) | NC score | 0.486844 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |
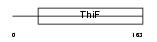
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.482618 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |
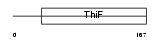
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULA1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.366413 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
ULA1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.361107 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ATG7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.145493 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
ATG7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.144726 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
KAT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.059900 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFU3, Q5SY48, Q5SY49, Q5SY50, Q5SY51, Q8NFU2, Q9BV22 | Gene names | KAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative thiosulfate sulfurtransferase KAT (EC 2.8.1.1). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.013671 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
ZFYV9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.014898 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.013919 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MOCS3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |

|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
ULE1B_MOUSE
|
||||||
| NC score | 0.669466 (rank : 2) | θ value | 2.88119e-14 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULE1B_HUMAN
|
||||||
| NC score | 0.643794 (rank : 3) | θ value | 1.9326e-10 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBE1L_HUMAN
|
||||||
| NC score | 0.617097 (rank : 4) | θ value | 2.52405e-10 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
UBA3_MOUSE
|
||||||
| NC score | 0.610921 (rank : 5) | θ value | 4.0297e-08 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBE1_MOUSE
|
||||||
| NC score | 0.609982 (rank : 6) | θ value | 6.64225e-11 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
UBA3_HUMAN
|
||||||
| NC score | 0.609670 (rank : 7) | θ value | 5.26297e-08 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBE1_HUMAN
|
||||||
| NC score | 0.604913 (rank : 8) | θ value | 9.59137e-10 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UE1D1_MOUSE
|
||||||
| NC score | 0.503647 (rank : 9) | θ value | 2.61198e-07 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
UE1D1_HUMAN
|
||||||
| NC score | 0.502997 (rank : 10) | θ value | 4.45536e-07 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
ULE1A_HUMAN
|
||||||
| NC score | 0.486844 (rank : 11) | θ value | 0.000461057 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_MOUSE
|
||||||
| NC score | 0.482618 (rank : 12) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULA1_MOUSE
|
||||||
| NC score | 0.366413 (rank : 13) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
ULA1_HUMAN
|
||||||
| NC score | 0.361107 (rank : 14) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ATG7_HUMAN
|
||||||
| NC score | 0.145493 (rank : 15) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
ATG7_MOUSE
|
||||||
| NC score | 0.144726 (rank : 16) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
KAT_HUMAN
|
||||||
| NC score | 0.059900 (rank : 17) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFU3, Q5SY48, Q5SY49, Q5SY50, Q5SY51, Q8NFU2, Q9BV22 | Gene names | KAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative thiosulfate sulfurtransferase KAT (EC 2.8.1.1). | |||||
|
ZFYV9_HUMAN
|
||||||
| NC score | 0.014898 (rank : 18) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.013919 (rank : 19) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.013671 (rank : 20) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||