Please be patient as the page loads
|
XRCC1_HUMAN
|
||||||
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
XRCC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.914098 (rank : 2) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
TRPC2_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 3) | NC score | 0.134812 (rank : 5) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 4) | NC score | 0.076404 (rank : 25) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
REV1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 5) | NC score | 0.131154 (rank : 6) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.127316 (rank : 7) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.032454 (rank : 132) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.070986 (rank : 37) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.119040 (rank : 9) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
TOPB1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.147604 (rank : 4) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.150710 (rank : 3) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.120099 (rank : 8) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.058758 (rank : 72) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.038751 (rank : 121) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.081559 (rank : 18) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
GOG8A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.036355 (rank : 124) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.052541 (rank : 99) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.074756 (rank : 28) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.040285 (rank : 118) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.073457 (rank : 31) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.057884 (rank : 74) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.075379 (rank : 26) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.079655 (rank : 21) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
WDR43_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.074170 (rank : 29) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.067470 (rank : 44) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PESC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.110061 (rank : 11) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.035925 (rank : 126) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.013212 (rank : 168) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.027897 (rank : 140) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.046748 (rank : 111) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.073825 (rank : 30) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
DNL3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.057167 (rank : 79) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.066865 (rank : 45) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.038147 (rank : 122) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.034301 (rank : 128) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.077510 (rank : 24) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.059771 (rank : 69) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.111606 (rank : 10) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.078845 (rank : 23) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.080448 (rank : 19) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.069838 (rank : 38) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.066832 (rank : 46) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.016615 (rank : 161) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.027540 (rank : 143) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.073203 (rank : 32) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.029816 (rank : 134) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.044531 (rank : 112) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
AL2S4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.055341 (rank : 86) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
AN32B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.053117 (rank : 95) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
ASPH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.066335 (rank : 48) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
BARD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.028125 (rank : 138) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
CLGN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.025443 (rank : 145) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |
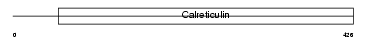
|
|||||
| Description | Calmegin precursor. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.072732 (rank : 33) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
FAM9A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.048872 (rank : 110) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
PAR10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.034027 (rank : 130) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
PESC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.104818 (rank : 12) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00541, Q6IC29 | Gene names | PES1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
PLCB3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.021232 (rank : 151) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
ZN592_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.014121 (rank : 166) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.014247 (rank : 165) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
NFL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.021017 (rank : 152) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.057698 (rank : 75) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
CCDC9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.052970 (rank : 96) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.014975 (rank : 163) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.015184 (rank : 162) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
DAF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.017317 (rank : 158) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.021418 (rank : 150) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.039106 (rank : 119) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.020851 (rank : 153) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.035004 (rank : 127) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TOR2A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.019572 (rank : 157) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1J9, Q3TBH0, Q3TC01, Q3V4D1, Q8R5B5 | Gene names | Tor2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-2A precursor (Torsin family 2 member A). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.011194 (rank : 170) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.090894 (rank : 15) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.063520 (rank : 56) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.007583 (rank : 177) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
THSD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.020543 (rank : 156) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.034046 (rank : 129) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.060952 (rank : 63) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.068292 (rank : 41) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
FETUB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.024815 (rank : 147) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXC1 | Gene names | Fetub | |||
|
Domain Architecture |
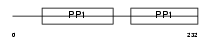
|
|||||
| Description | Fetuin-B precursor (IRL685). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.032456 (rank : 131) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.020628 (rank : 155) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.010545 (rank : 172) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.058062 (rank : 73) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.065127 (rank : 52) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.059060 (rank : 71) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.080196 (rank : 20) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.009652 (rank : 175) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.009878 (rank : 173) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
ULE1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.030820 (rank : 133) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ZHX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.016867 (rank : 159) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
BIM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.024875 (rank : 146) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
CEP41_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.022233 (rank : 149) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.013017 (rank : 169) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.026160 (rank : 144) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.009525 (rank : 176) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.028364 (rank : 137) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.010663 (rank : 171) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.064778 (rank : 53) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.014774 (rank : 164) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.050596 (rank : 106) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.027774 (rank : 141) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
STAG3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.020656 (rank : 154) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJ98, Q8NDP3 | Gene names | STAG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
TNR8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.027642 (rank : 142) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |
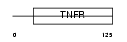
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
AN32E_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.041641 (rank : 115) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
BCL6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.001966 (rank : 181) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |
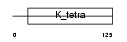
|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.022467 (rank : 148) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
DRAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.028086 (rank : 139) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.049382 (rank : 107) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.038997 (rank : 120) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.006736 (rank : 178) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.037398 (rank : 123) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PO3F4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.006534 (rank : 179) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62515, P25216 | Gene names | Pou3f4, Brn-4, Brn4, Otf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 3, transcription factor 4 (Brain-specific homeobox/POU domain protein 4) (Brain-4) (Brn-4). | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.029235 (rank : 136) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.029552 (rank : 135) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
UT14A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.016735 (rank : 160) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
ZSCA5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.001744 (rank : 182) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
AIOL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.001335 (rank : 183) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08900 | Gene names | Ikzf3, Zfpn1a3, Znfn1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.079236 (rank : 22) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CTDP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.060491 (rank : 65) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.009819 (rank : 174) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.049353 (rank : 108) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.040469 (rank : 116) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.072146 (rank : 34) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.049025 (rank : 109) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.043346 (rank : 114) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SH3BG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.035925 (rank : 125) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.083649 (rank : 17) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TRI16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.006331 (rank : 180) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.040344 (rank : 117) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.044085 (rank : 113) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.013693 (rank : 167) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.060582 (rank : 64) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
AN32A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051811 (rank : 101) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.089898 (rank : 16) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.099884 (rank : 13) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.057133 (rank : 80) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.062041 (rank : 62) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.051854 (rank : 100) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.066394 (rank : 47) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.050657 (rank : 105) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.060177 (rank : 67) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
KI67_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.056185 (rank : 83) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.054874 (rank : 88) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.069119 (rank : 39) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.062790 (rank : 58) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.054125 (rank : 93) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.057447 (rank : 78) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.063609 (rank : 55) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.062350 (rank : 61) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.054319 (rank : 91) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.065476 (rank : 50) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.051187 (rank : 103) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.068048 (rank : 42) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.055676 (rank : 85) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.060304 (rank : 66) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.055121 (rank : 87) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.055874 (rank : 84) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.057057 (rank : 81) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.050825 (rank : 104) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.067888 (rank : 43) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.063753 (rank : 54) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.054705 (rank : 90) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.057019 (rank : 82) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.099417 (rank : 14) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
REV1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.057451 (rank : 77) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.062405 (rank : 60) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.052557 (rank : 98) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.068840 (rank : 40) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.071888 (rank : 35) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.062558 (rank : 59) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.071329 (rank : 36) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.054213 (rank : 92) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.059559 (rank : 70) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.075125 (rank : 27) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.060121 (rank : 68) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.063026 (rank : 57) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.053211 (rank : 94) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H322, Q9P0H5 | Gene names | VCX2, VCX2R, VCXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 2 (VCX-B protein) (Variably charged protein X-B) (Variable charge protein on X with two repeats) (VCX-2r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.066080 (rank : 49) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.065166 (rank : 51) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
VCY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.054856 (rank : 89) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
VGF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.057618 (rank : 76) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.052584 (rank : 97) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.051484 (rank : 102) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
XRCC1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.914098 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.150710 (rank : 3) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
TOPB1_HUMAN
|
||||||
| NC score | 0.147604 (rank : 4) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.134812 (rank : 5) | θ value | 6.87365e-08 (rank : 3) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
REV1_MOUSE
|
||||||
| NC score | 0.131154 (rank : 6) | θ value | 0.000461057 (rank : 5) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.127316 (rank : 7) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.120099 (rank : 8) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.119040 (rank : 9) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.111606 (rank : 10) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PESC_MOUSE
|
||||||
| NC score | 0.110061 (rank : 11) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
PESC_HUMAN
|
||||||
| NC score | 0.104818 (rank : 12) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00541, Q6IC29 | Gene names | PES1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.099884 (rank : 13) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.099417 (rank : 14) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.090894 (rank : 15) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.089898 (rank : 16) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.083649 (rank : 17) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.081559 (rank : 18) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.080448 (rank : 19) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.080196 (rank : 20) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.079655 (rank : 21) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.079236 (rank : 22) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.078845 (rank : 23) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.077510 (rank : 24) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.076404 (rank : 25) | θ value | 1.09739e-05 (rank : 4) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.075379 (rank : 26) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.075125 (rank : 27) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.074756 (rank : 28) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
WDR43_HUMAN
|
||||||
| NC score | 0.074170 (rank : 29) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.073825 (rank : 30) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.073457 (rank : 31) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.073203 (rank : 32) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.072732 (rank : 33) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.072146 (rank : 34) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.071888 (rank : 35) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.071329 (rank : 36) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.070986 (rank : 37) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.069838 (rank : 38) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.069119 (rank : 39) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.068840 (rank : 40) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.068292 (rank : 41) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.068048 (rank : 42) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.067888 (rank : 43) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.067470 (rank : 44) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.066865 (rank : 45) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.066832 (rank : 46) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.066394 (rank : 47) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ASPH_HUMAN
|
||||||
| NC score | 0.066335 (rank : 48) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.066080 (rank : 49) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.065476 (rank : 50) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.065166 (rank : 51) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.065127 (rank : 52) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.064778 (rank : 53) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.063753 (rank : 54) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.063609 (rank : 55) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.063520 (rank : 56) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.063026 (rank : 57) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.062790 (rank : 58) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.062558 (rank : 59) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.062405 (rank : 60) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.062350 (rank : 61) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.062041 (rank : 62) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.060952 (rank : 63) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.060582 (rank : 64) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CTDP1_MOUSE
|
||||||
| NC score | 0.060491 (rank : 65) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.060304 (rank : 66) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.060177 (rank : 67) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.060121 (rank : 68) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.059771 (rank : 69) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.059559 (rank : 70) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.059060 (rank : 71) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.058758 (rank : 72) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.058062 (rank : 73) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.057884 (rank : 74) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.057698 (rank : 75) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
VGF_HUMAN
|
||||||
| NC score | 0.057618 (rank : 76) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
REV1_HUMAN
|
||||||
| NC score | 0.057451 (rank : 77) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.057447 (rank : 78) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
DNL3_HUMAN
|
||||||
| NC score | 0.057167 (rank : 79) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.057133 (rank : 80) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.057057 (rank : 81) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.057019 (rank : 82) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.056185 (rank : 83) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.055874 (rank : 84) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.055676 (rank : 85) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
AL2S4_MOUSE
|
||||||
| NC score | 0.055341 (rank : 86) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.055121 (rank : 87) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.054874 (rank : 88) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
VCY1_HUMAN
|
||||||
| NC score | 0.054856 (rank : 89) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.054705 (rank : 90) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.054319 (rank : 91) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.054213 (rank : 92) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.054125 (rank : 93) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
VCX2_HUMAN
|
||||||
| NC score | 0.053211 (rank : 94) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H322, Q9P0H5 | Gene names | VCX2, VCX2R, VCXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 2 (VCX-B protein) (Variably charged protein X-B) (Variable charge protein on X with two repeats) (VCX-2r). | |||||
|
AN32B_HUMAN
|
||||||
| NC score | 0.053117 (rank : 95) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
CCDC9_HUMAN
|
||||||
| NC score | 0.052970 (rank : 96) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.052584 (rank : 97) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.052557 (rank : 98) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.052541 (rank : 99) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.051854 (rank : 100) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
AN32A_HUMAN
|
||||||
| NC score | 0.051811 (rank : 101) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.051484 (rank : 102) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.051187 (rank : 103) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.050825 (rank : 104) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.050657 (rank : 105) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.050596 (rank : 106) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.049382 (rank : 107) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.049353 (rank : 108) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.049025 (rank : 109) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
FAM9A_HUMAN
|
||||||
| NC score | 0.048872 (rank : 110) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.046748 (rank : 111) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.044531 (rank : 112) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.044085 (rank : 113) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.043346 (rank : 114) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
AN32E_HUMAN
|
||||||
| NC score | 0.041641 (rank : 115) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.040469 (rank : 116) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.040344 (rank : 117) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.040285 (rank : 118) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.039106 (rank : 119) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.038997 (rank : 120) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.038751 (rank : 121) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.038147 (rank : 122) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.037398 (rank : 123) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
GOG8A_HUMAN
|
||||||
| NC score | 0.036355 (rank : 124) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
SH3BG_MOUSE
|
||||||
| NC score | 0.035925 (rank : 125) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.035925 (rank : 126) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.035004 (rank : 127) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.034301 (rank : 128) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.034046 (rank : 129) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
PAR10_HUMAN
|
||||||
| NC score | 0.034027 (rank : 130) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.032456 (rank : 131) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.032454 (rank : 132) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ULE1B_MOUSE
|
||||||
| NC score | 0.030820 (rank : 133) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.029816 (rank : 134) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.029552 (rank : 135) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.029235 (rank : 136) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.028364 (rank : 137) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
BARD1_HUMAN
|
||||||
| NC score | 0.028125 (rank : 138) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
DRAP1_MOUSE
|
||||||
| NC score | 0.028086 (rank : 139) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.027897 (rank : 140) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.027774 (rank : 141) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TNR8_MOUSE
|
||||||
| NC score | 0.027642 (rank : 142) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.027540 (rank : 143) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.026160 (rank : 144) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
CLGN_HUMAN
|
||||||
| NC score | 0.025443 (rank : 145) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor. | |||||
|
BIM_HUMAN
|
||||||
| NC score | 0.024875 (rank : 146) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
FETUB_MOUSE
|
||||||
| NC score | 0.024815 (rank : 147) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXC1 | Gene names | Fetub | |||
|
Domain Architecture |

|
|||||
| Description | Fetuin-B precursor (IRL685). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.022467 (rank : 148) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
CEP41_HUMAN
|
||||||
| NC score | 0.022233 (rank : 149) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.021418 (rank : 150) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
PLCB3_MOUSE
|
||||||
| NC score | 0.021232 (rank : 151) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.021017 (rank : 152) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.020851 (rank : 153) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
STAG3_HUMAN
|
||||||
| NC score | 0.020656 (rank : 154) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJ98, Q8NDP3 | Gene names | STAG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.020628 (rank : 155) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
THSD1_HUMAN
|
||||||
| NC score | 0.020543 (rank : 156) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
TOR2A_MOUSE
|
||||||
| NC score | 0.019572 (rank : 157) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1J9, Q3TBH0, Q3TC01, Q3V4D1, Q8R5B5 | Gene names | Tor2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-2A precursor (Torsin family 2 member A). | |||||
|
DAF1_MOUSE
|
||||||
| NC score | 0.017317 (rank : 158) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
ZHX1_MOUSE
|
||||||
| NC score | 0.016867 (rank : 159) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
UT14A_MOUSE
|
||||||
| NC score | 0.016735 (rank : 160) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.016615 (rank : 161) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.015184 (rank : 162) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.014975 (rank : 163) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.014774 (rank : 164) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.014247 (rank : 165) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
ZN592_MOUSE
|
||||||
| NC score | 0.014121 (rank : 166) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.013693 (rank : 167) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.013212 (rank : 168) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.013017 (rank : 169) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.011194 (rank : 170) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.010663 (rank : 171) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.010545 (rank : 172) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.009878 (rank : 173) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.009819 (rank : 174) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.009652 (rank : 175) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.009525 (rank : 176) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | 0.007583 (rank : 177) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.006736 (rank : 178) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PO3F4_MOUSE
|
||||||
| NC score | 0.006534 (rank : 179) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62515, P25216 | Gene names | Pou3f4, Brn-4, Brn4, Otf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 3, transcription factor 4 (Brain-specific homeobox/POU domain protein 4) (Brain-4) (Brn-4). | |||||
|
TRI16_HUMAN
|
||||||
| NC score | 0.006331 (rank : 180) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
BCL6_HUMAN
|
||||||
| NC score | 0.001966 (rank : 181) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||
|
ZSCA5_HUMAN
|
||||||
| NC score | 0.001744 (rank : 182) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
AIOL_MOUSE
|
||||||
| NC score | 0.001335 (rank : 183) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08900 | Gene names | Ikzf3, Zfpn1a3, Znfn1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||