Please be patient as the page loads
|
MAGB6_HUMAN
|
||||||
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MAGB6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 184 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MAGB4_HUMAN
|
||||||
| θ value | 4.37348e-71 (rank : 2) | NC score | 0.978195 (rank : 2) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGB1_HUMAN
|
||||||
| θ value | 1.27248e-70 (rank : 3) | NC score | 0.977702 (rank : 3) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MAGB2_HUMAN
|
||||||
| θ value | 2.17557e-62 (rank : 4) | NC score | 0.977349 (rank : 4) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
MAGB3_HUMAN
|
||||||
| θ value | 1.55911e-60 (rank : 5) | NC score | 0.975785 (rank : 6) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
MAGBI_HUMAN
|
||||||
| θ value | 2.25136e-59 (rank : 6) | NC score | 0.975136 (rank : 7) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
MAGAA_HUMAN
|
||||||
| θ value | 2.94036e-59 (rank : 7) | NC score | 0.967558 (rank : 8) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
MAGBA_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 8) | NC score | 0.976586 (rank : 5) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGAB_HUMAN
|
||||||
| θ value | 5.19021e-56 (rank : 9) | NC score | 0.963937 (rank : 11) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGA9_HUMAN
|
||||||
| θ value | 2.41096e-53 (rank : 10) | NC score | 0.964395 (rank : 10) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
MAGA4_HUMAN
|
||||||
| θ value | 4.54686e-52 (rank : 11) | NC score | 0.955796 (rank : 16) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
MAGA1_HUMAN
|
||||||
| θ value | 8.57503e-51 (rank : 12) | NC score | 0.958199 (rank : 15) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |
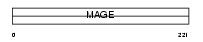
|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGG1_HUMAN
|
||||||
| θ value | 3.72821e-46 (rank : 13) | NC score | 0.963865 (rank : 12) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |
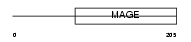
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 1.19932e-44 (rank : 14) | NC score | 0.906429 (rank : 34) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAGA2_HUMAN
|
||||||
| θ value | 7.77379e-44 (rank : 15) | NC score | 0.947164 (rank : 18) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |
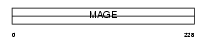
|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGAC_HUMAN
|
||||||
| θ value | 5.03882e-43 (rank : 16) | NC score | 0.946323 (rank : 19) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |
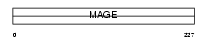
|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGA6_HUMAN
|
||||||
| θ value | 1.46607e-42 (rank : 17) | NC score | 0.946276 (rank : 20) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MAGA3_HUMAN
|
||||||
| θ value | 1.91475e-42 (rank : 18) | NC score | 0.945256 (rank : 21) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGC2_HUMAN
|
||||||
| θ value | 3.26607e-42 (rank : 19) | NC score | 0.966688 (rank : 9) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
MAGG1_MOUSE
|
||||||
| θ value | 7.27602e-42 (rank : 20) | NC score | 0.962491 (rank : 13) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 1.2411e-41 (rank : 21) | NC score | 0.672136 (rank : 36) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | 1.16164e-39 (rank : 22) | NC score | 0.947618 (rank : 17) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
TROP_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 23) | NC score | 0.904137 (rank : 35) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
MAGF1_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 24) | NC score | 0.961525 (rank : 14) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 25) | NC score | 0.907639 (rank : 33) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
MAGL2_HUMAN
|
||||||
| θ value | 1.42001e-37 (rank : 26) | NC score | 0.942884 (rank : 22) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
MAGD1_MOUSE
|
||||||
| θ value | 2.67802e-36 (rank : 27) | NC score | 0.919243 (rank : 31) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAGD1_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 28) | NC score | 0.925420 (rank : 27) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MAGL2_MOUSE
|
||||||
| θ value | 2.19584e-30 (rank : 29) | NC score | 0.933321 (rank : 25) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |
|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
NECD_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 30) | NC score | 0.934157 (rank : 23) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |
|
|||||
| Description | Necdin. | |||||
|
MAGA8_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 31) | NC score | 0.922994 (rank : 28) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
NECD_MOUSE
|
||||||
| θ value | 1.08979e-29 (rank : 32) | NC score | 0.929129 (rank : 26) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MAGE2_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 33) | NC score | 0.933745 (rank : 24) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
MAGC3_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 34) | NC score | 0.907753 (rank : 32) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
MAGH1_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 35) | NC score | 0.922968 (rank : 29) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
MAGH1_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 36) | NC score | 0.921268 (rank : 30) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 37) | NC score | 0.059560 (rank : 48) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 38) | NC score | 0.060746 (rank : 47) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
BASP_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 39) | NC score | 0.090792 (rank : 38) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 40) | NC score | 0.046789 (rank : 57) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 41) | NC score | 0.026571 (rank : 97) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 42) | NC score | 0.065139 (rank : 41) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 43) | NC score | 0.064440 (rank : 44) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BCAS1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.046640 (rank : 58) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
MAGA5_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 45) | NC score | 0.597831 (rank : 37) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |
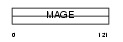
|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
SFRS7_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 46) | NC score | 0.018998 (rank : 115) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
LBXCO_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 47) | NC score | 0.033184 (rank : 75) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
I15RA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.037360 (rank : 69) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13261, Q6B0J2, Q7LDR4, Q7Z609 | Gene names | IL15RA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-15 receptor alpha chain precursor (IL-15R-alpha) (IL- 15RA). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.125558 (rank : 49) | NC score | 0.064901 (rank : 43) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 50) | NC score | 0.026959 (rank : 96) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 0.163984 (rank : 51) | NC score | 0.077908 (rank : 39) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
NFAC1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 52) | NC score | 0.028823 (rank : 87) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.163984 (rank : 53) | NC score | 0.009788 (rank : 146) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 54) | NC score | 0.039733 (rank : 68) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.007525 (rank : 154) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
CA106_MOUSE
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.030428 (rank : 80) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.21417 (rank : 57) | NC score | 0.041642 (rank : 63) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
REXO1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 58) | NC score | 0.029141 (rank : 84) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 59) | NC score | 0.077177 (rank : 40) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 60) | NC score | 0.035913 (rank : 71) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CTDP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 61) | NC score | 0.028235 (rank : 90) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
TNR1B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 62) | NC score | 0.015258 (rank : 127) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |
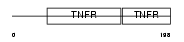
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 0.365318 (rank : 63) | NC score | 0.029708 (rank : 82) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 64) | NC score | 0.045060 (rank : 62) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PAK4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 65) | NC score | 0.000582 (rank : 177) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O96013, Q8N4E1, Q8NCH5, Q8NDE3, Q9BU33, Q9ULS8 | Gene names | PAK4, KIAA1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 66) | NC score | 0.063437 (rank : 45) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.029381 (rank : 83) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 68) | NC score | 0.032777 (rank : 77) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.47712 (rank : 69) | NC score | 0.017560 (rank : 118) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
NF2L3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 70) | NC score | 0.015573 (rank : 126) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.021896 (rank : 105) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.033091 (rank : 76) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.053412 (rank : 52) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 74) | NC score | 0.004874 (rank : 171) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.041097 (rank : 66) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.011802 (rank : 141) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.016989 (rank : 120) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.026334 (rank : 98) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.062010 (rank : 46) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.034483 (rank : 73) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TCEA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.017363 (rank : 119) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10711, P10712, P23713 | Gene names | Tcea1, Tceat | |||
|
Domain Architecture |
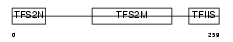
|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.020762 (rank : 108) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.046492 (rank : 59) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.027802 (rank : 93) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.028931 (rank : 86) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | 0.046018 (rank : 61) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | 0.041528 (rank : 64) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 88) | NC score | 0.008995 (rank : 151) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
DCP1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.014746 (rank : 131) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.029051 (rank : 85) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
RANB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.027756 (rank : 94) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H6Z4, O60405, O75759, O75760, Q9BT47, Q9UG74 | Gene names | RANBP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.028200 (rank : 91) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TCEA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.015714 (rank : 125) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |
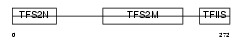
|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.005417 (rank : 166) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.001783 (rank : 176) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.033194 (rank : 74) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.027539 (rank : 95) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.005321 (rank : 168) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
BCL7C_MOUSE
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.014054 (rank : 133) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
MARCO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.004430 (rank : 172) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.014878 (rank : 128) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.012247 (rank : 138) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.016639 (rank : 121) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANR38_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.005438 (rank : 165) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T7N3, Q6P9A0, Q86T71, Q86VE6, Q8NAX3 | Gene names | ANKRD38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.020722 (rank : 109) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.006008 (rank : 162) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
HCLS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.009291 (rank : 149) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
JIP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.009754 (rank : 147) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.005505 (rank : 164) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.028339 (rank : 89) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NOL4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.011863 (rank : 140) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60954 | Gene names | Nol4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 4. | |||||
|
SFR16_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.018943 (rank : 116) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.022552 (rank : 102) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
APC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.028141 (rank : 92) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
CADH6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.000396 (rank : 178) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55285, Q9BWS0 | Gene names | CDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.016411 (rank : 123) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.009194 (rank : 150) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CLAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.012839 (rank : 134) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.018330 (rank : 117) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | -0.003391 (rank : 187) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.037247 (rank : 70) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.012050 (rank : 139) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PITX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | -0.001051 (rank : 184) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78337, O14677, O60425, Q9BTI5 | Gene names | PITX1, BFT, PTX1 | |||
|
Domain Architecture |
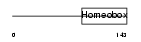
|
|||||
| Description | Pituitary homeobox 1 (Hindlimb expressed homeobox protein backfoot). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.047655 (rank : 56) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.028606 (rank : 88) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
XRCC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.020628 (rank : 110) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
APC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.030217 (rank : 81) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.006643 (rank : 159) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
CADH6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.000295 (rank : 179) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97326, P70393 | Gene names | Cdh6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.005593 (rank : 163) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CLAP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.014877 (rank : 129) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75122, Q7L8F6, Q8N6R6, Q9BQT3, Q9BQT4, Q9H7A3, Q9NSZ2 | Gene names | CLASP2, KIAA0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CO1A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.012659 (rank : 135) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.014464 (rank : 132) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.034842 (rank : 72) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.051803 (rank : 54) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SEN34_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.019009 (rank : 114) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.032547 (rank : 78) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.056988 (rank : 49) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TNT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.015886 (rank : 124) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2V1, Q8NEF0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TNT. | |||||
|
TUB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.006271 (rank : 160) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50607, O00293 | Gene names | TUB | |||
|
Domain Architecture |

|
|||||
| Description | Tubby protein homolog. | |||||
|
UBN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.030725 (rank : 79) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.003414 (rank : 174) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.019673 (rank : 111) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
BRAC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.002207 (rank : 175) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15178 | Gene names | T | |||
|
Domain Architecture |
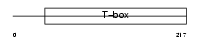
|
|||||
| Description | Brachyury protein (T protein). | |||||
|
CDSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.022220 (rank : 104) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.007452 (rank : 155) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.011772 (rank : 142) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.016604 (rank : 122) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
K0310_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.021014 (rank : 107) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
LRRK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | -0.003985 (rank : 188) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHC2, Q3U476, Q66JQ4, Q6GQR9, Q6NZF5, Q6ZPI4, Q8BKP3, Q8BU93, Q8BUY0, Q8BVV2, Q8R085 | Gene names | Lrrk1, Kiaa1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||
|
PACS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.024792 (rank : 99) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6VY07, Q6PJY6, Q6PKB6, Q7Z590, Q7Z5W4, Q8N8K6, Q96MW0, Q9NW92, Q9ULP5 | Gene names | PACS1, KIAA1175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
PACS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.022403 (rank : 103) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
RAE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.005394 (rank : 167) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.021683 (rank : 106) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SNX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.003474 (rank : 173) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWK8, Q3UKW3, Q9CQV0 | Gene names | Snx2 | |||
|
Domain Architecture |
|
|||||
| Description | Sorting nexin-2. | |||||
|
SPHK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.006731 (rank : 157) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NRA0, Q9BRN1, Q9H0Q2, Q9NWU7 | Gene names | SPHK2 | |||
|
Domain Architecture |
|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
ST5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.008747 (rank : 152) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.040077 (rank : 67) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
TOPB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.008123 (rank : 153) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | -0.000339 (rank : 180) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.046257 (rank : 60) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.041327 (rank : 65) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.012635 (rank : 136) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
ASCC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.010150 (rank : 145) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H1I8, Q4TT54, Q8TAZ0, Q9H711, Q9H9D6 | Gene names | ASCC2, ASC1P100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 2 (ASC-1 complex subunit p100) (Trip4 complex subunit p100). | |||||
|
CAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.004981 (rank : 170) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |
|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CE016_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.006677 (rank : 158) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
CE152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | -0.000622 (rank : 183) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.007342 (rank : 156) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.010944 (rank : 143) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.023031 (rank : 101) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.023936 (rank : 100) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | -0.002034 (rank : 185) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
M3KL4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | -0.003083 (rank : 186) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5TCX8, Q5TCX7, Q5TCX9, Q8WWN1, Q8WWN2, Q96JM1 | Gene names | MLK4, KIAA1804 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase (EC 2.7.11.25) (Mixed lineage kinase 4). | |||||
|
MYCD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.012489 (rank : 137) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | -0.000396 (rank : 182) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.019333 (rank : 113) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.014837 (rank : 130) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
POLS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.005107 (rank : 169) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | -0.000345 (rank : 181) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.052849 (rank : 53) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TYDP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.006229 (rank : 161) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ37, Q8VE39 | Gene names | Tdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosyl-DNA phosphodiesterase 1 (EC 3.1.4.-) (Tyr-DNA phosphodiesterase 1). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 182) | NC score | 0.009360 (rank : 148) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 8.99809 (rank : 183) | NC score | 0.010344 (rank : 144) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 8.99809 (rank : 184) | NC score | 0.019521 (rank : 112) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.056321 (rank : 50) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
LORI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.065100 (rank : 42) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
PHXR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.055307 (rank : 51) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
SELPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.050263 (rank : 55) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 184 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MAGB4_HUMAN
|
||||||
| NC score | 0.978195 (rank : 2) | θ value | 4.37348e-71 (rank : 2) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGB1_HUMAN
|
||||||
| NC score | 0.977702 (rank : 3) | θ value | 1.27248e-70 (rank : 3) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MAGB2_HUMAN
|
||||||
| NC score | 0.977349 (rank : 4) | θ value | 2.17557e-62 (rank : 4) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
MAGBA_HUMAN
|
||||||
| NC score | 0.976586 (rank : 5) | θ value | 2.48917e-58 (rank : 8) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGB3_HUMAN
|
||||||
| NC score | 0.975785 (rank : 6) | θ value | 1.55911e-60 (rank : 5) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
MAGBI_HUMAN
|
||||||
| NC score | 0.975136 (rank : 7) | θ value | 2.25136e-59 (rank : 6) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
MAGAA_HUMAN
|
||||||
| NC score | 0.967558 (rank : 8) | θ value | 2.94036e-59 (rank : 7) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
MAGC2_HUMAN
|
||||||
| NC score | 0.966688 (rank : 9) | θ value | 3.26607e-42 (rank : 19) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
MAGA9_HUMAN
|
||||||
| NC score | 0.964395 (rank : 10) | θ value | 2.41096e-53 (rank : 10) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
MAGAB_HUMAN
|
||||||
| NC score | 0.963937 (rank : 11) | θ value | 5.19021e-56 (rank : 9) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGG1_HUMAN
|
||||||
| NC score | 0.963865 (rank : 12) | θ value | 3.72821e-46 (rank : 13) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGG1_MOUSE
|
||||||
| NC score | 0.962491 (rank : 13) | θ value | 7.27602e-42 (rank : 20) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MAGF1_HUMAN
|
||||||
| NC score | 0.961525 (rank : 14) | θ value | 2.86122e-38 (rank : 24) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGA1_HUMAN
|
||||||
| NC score | 0.958199 (rank : 15) | θ value | 8.57503e-51 (rank : 12) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGA4_HUMAN
|
||||||
| NC score | 0.955796 (rank : 16) | θ value | 4.54686e-52 (rank : 11) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.947618 (rank : 17) | θ value | 1.16164e-39 (rank : 22) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MAGA2_HUMAN
|
||||||
| NC score | 0.947164 (rank : 18) | θ value | 7.77379e-44 (rank : 15) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGAC_HUMAN
|
||||||
| NC score | 0.946323 (rank : 19) | θ value | 5.03882e-43 (rank : 16) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGA6_HUMAN
|
||||||
| NC score | 0.946276 (rank : 20) | θ value | 1.46607e-42 (rank : 17) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MAGA3_HUMAN
|
||||||
| NC score | 0.945256 (rank : 21) | θ value | 1.91475e-42 (rank : 18) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGL2_HUMAN
|
||||||
| NC score | 0.942884 (rank : 22) | θ value | 1.42001e-37 (rank : 26) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
NECD_HUMAN
|
||||||
| NC score | 0.934157 (rank : 23) | θ value | 2.19584e-30 (rank : 30) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MAGE2_HUMAN
|
||||||
| NC score | 0.933745 (rank : 24) | θ value | 2.42779e-29 (rank : 33) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
MAGL2_MOUSE
|
||||||
| NC score | 0.933321 (rank : 25) | θ value | 2.19584e-30 (rank : 29) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
NECD_MOUSE
|
||||||
| NC score | 0.929129 (rank : 26) | θ value | 1.08979e-29 (rank : 32) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.925420 (rank : 27) | θ value | 5.96599e-36 (rank : 28) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MAGA8_HUMAN
|
||||||
| NC score | 0.922994 (rank : 28) | θ value | 1.08979e-29 (rank : 31) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
MAGH1_HUMAN
|
||||||
| NC score | 0.922968 (rank : 29) | θ value | 1.42661e-21 (rank : 35) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
MAGH1_MOUSE
|
||||||
| NC score | 0.921268 (rank : 30) | θ value | 1.86321e-21 (rank : 36) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
MAGD1_MOUSE
|
||||||
| NC score | 0.919243 (rank : 31) | θ value | 2.67802e-36 (rank : 27) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAGC3_HUMAN
|
||||||
| NC score | 0.907753 (rank : 32) | θ value | 6.84181e-24 (rank : 34) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.907639 (rank : 33) | θ value | 4.88048e-38 (rank : 25) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.906429 (rank : 34) | θ value | 1.19932e-44 (rank : 14) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
TROP_HUMAN
|
||||||
| NC score | 0.904137 (rank : 35) | θ value | 1.98146e-39 (rank : 23) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.672136 (rank : 36) | θ value | 1.2411e-41 (rank : 21) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGA5_HUMAN
|
||||||
| NC score | 0.597831 (rank : 37) | θ value | 0.0736092 (rank : 45) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.090792 (rank : 38) | θ value | 0.00869519 (rank : 39) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.077908 (rank : 39) | θ value | 0.163984 (rank : 51) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.077177 (rank : 40) | θ value | 0.21417 (rank : 59) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.065139 (rank : 41) | θ value | 0.0330416 (rank : 42) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.065100 (rank : 42) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.064901 (rank : 43) | θ value | 0.125558 (rank : 49) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.064440 (rank : 44) | θ value | 0.0330416 (rank : 43) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.063437 (rank : 45) | θ value | 0.365318 (rank : 66) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.062010 (rank : 46) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.060746 (rank : 47) | θ value | 0.00298849 (rank : 38) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.059560 (rank : 48) | θ value | 0.00020696 (rank : 37) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.056988 (rank : 49) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.056321 (rank : 50) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
PHXR5_MOUSE
|
||||||
| NC score | 0.055307 (rank : 51) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.053412 (rank : 52) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.052849 (rank : 53) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.051803 (rank : 54) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.050263 (rank : 55) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.047655 (rank : 56) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.046789 (rank : 57) | θ value | 0.0193708 (rank : 40) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
BCAS1_HUMAN
|
||||||
| NC score | 0.046640 (rank : 58) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.046492 (rank : 59) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.046257 (rank : 60) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.046018 (rank : 61) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.045060 (rank : 62) | θ value | 0.365318 (rank : 64) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.041642 (rank : 63) | θ value | 0.21417 (rank : 57) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.041528 (rank : 64) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.041327 (rank : 65) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.041097 (rank : 66) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.040077 (rank : 67) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.039733 (rank : 68) | θ value | 0.163984 (rank : 54) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
I15RA_HUMAN
|
||||||
| NC score | 0.037360 (rank : 69) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13261, Q6B0J2, Q7LDR4, Q7Z609 | Gene names | IL15RA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-15 receptor alpha chain precursor (IL-15R-alpha) (IL- 15RA). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.037247 (rank : 70) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.035913 (rank : 71) | θ value | 0.279714 (rank : 60) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.034842 (rank : 72) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.034483 (rank : 73) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.033194 (rank : 74) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
LBXCO_HUMAN
|
||||||
| NC score | 0.033184 (rank : 75) | θ value | 0.0961366 (rank : 47) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.033091 (rank : 76) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.032777 (rank : 77) | θ value | 0.47712 (rank : 68) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.032547 (rank : 78) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
UBN1_HUMAN
|
||||||
| NC score | 0.030725 (rank : 79) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
CA106_MOUSE
|
||||||
| NC score | 0.030428 (rank : 80) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.030217 (rank : 81) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.029708 (rank : 82) | θ value | 0.365318 (rank : 63) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.029381 (rank : 83) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
REXO1_MOUSE
|
||||||
| NC score | 0.029141 (rank : 84) | θ value | 0.21417 (rank : 58) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.029051 (rank : 85) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.028931 (rank : 86) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
NFAC1_MOUSE
|
||||||
| NC score | 0.028823 (rank : 87) | θ value | 0.163984 (rank : 52) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.028606 (rank : 88) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.028339 (rank : 89) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CTDP1_MOUSE
|
||||||
| NC score | 0.028235 (rank : 90) | θ value | 0.279714 (rank : 61) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.028200 (rank : 91) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.028141 (rank : 92) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.027802 (rank : 93) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
RANB3_HUMAN
|
||||||
| NC score | 0.027756 (rank : 94) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H6Z4, O60405, O75759, O75760, Q9BT47, Q9UG74 | Gene names | RANBP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.027539 (rank : 95) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.026959 (rank : 96) | θ value | 0.125558 (rank : 50) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.026571 (rank : 97) | θ value | 0.0252991 (rank : 41) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.026334 (rank : 98) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PACS1_HUMAN
|
||||||
| NC score | 0.024792 (rank : 99) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6VY07, Q6PJY6, Q6PKB6, Q7Z590, Q7Z5W4, Q8N8K6, Q96MW0, Q9NW92, Q9ULP5 | Gene names | PACS1, KIAA1175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.023936 (rank : 100) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.023031 (rank : 101) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.022552 (rank : 102) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
PACS1_MOUSE
|
||||||
| NC score | 0.022403 (rank : 103) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.022220 (rank : 104) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.021896 (rank : 105) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.021683 (rank : 106) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.021014 (rank : 107) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.020762 (rank : 108) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.020722 (rank : 109) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
XRCC1_HUMAN
|
||||||
| NC score | 0.020628 (rank : 110) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.019673 (rank : 111) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.019521 (rank : 112) | θ value | 8.99809 (rank : 184) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.019333 (rank : 113) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
SEN34_MOUSE
|
||||||
| NC score | 0.019009 (rank : 114) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
SFRS7_MOUSE
|
||||||
| NC score | 0.018998 (rank : 115) | θ value | 0.0736092 (rank : 46) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
SFR16_MOUSE
|
||||||
| NC score | 0.018943 (rank : 116) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.018330 (rank : 117) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.017560 (rank : 118) | θ value | 0.47712 (rank : 69) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
TCEA1_MOUSE
|
||||||
| NC score | 0.017363 (rank : 119) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10711, P10712, P23713 | Gene names | Tcea1, Tceat | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.016989 (rank : 120) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.016639 (rank : 121) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.016604 (rank : 122) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.016411 (rank : 123) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
TNT_HUMAN
|
||||||
| NC score | 0.015886 (rank : 124) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2V1, Q8NEF0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TNT. | |||||
|
TCEA1_HUMAN
|
||||||
| NC score | 0.015714 (rank : 125) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
NF2L3_MOUSE
|
||||||
| NC score | 0.015573 (rank : 126) | θ value | 0.47712 (rank : 70) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
TNR1B_MOUSE
|
||||||
| NC score | 0.015258 (rank : 127) | θ value | 0.279714 (rank : 62) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.014878 (rank : 128) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
CLAP2_HUMAN
|
||||||
| NC score | 0.014877 (rank : 129) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75122, Q7L8F6, Q8N6R6, Q9BQT3, Q9BQT4, Q9H7A3, Q9NSZ2 | Gene names | CLASP2, KIAA0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.014837 (rank : 130) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
DCP1A_MOUSE
|
||||||
| NC score | 0.014746 (rank : 131) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.014464 (rank : 132) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
BCL7C_MOUSE
|
||||||
| NC score | 0.014054 (rank : 133) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
CLAP2_MOUSE
|
||||||
| NC score | 0.012839 (rank : 134) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CO1A1_MOUSE
|
||||||
| NC score | 0.012659 (rank : 135) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.012635 (rank : 136) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
MYCD_HUMAN
|
||||||
| NC score | 0.012489 (rank : 137) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.012247 (rank : 138) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.012050 (rank : 139) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
NOL4_MOUSE
|
||||||
| NC score | 0.011863 (rank : 140) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60954 | Gene names | Nol4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 4. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.011802 (rank : 141) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.011772 (rank : 142) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.010944 (rank : 143) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.010344 (rank : 144) | θ value | 8.99809 (rank : 183) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
ASCC2_HUMAN
|
||||||
| NC score | 0.010150 (rank : 145) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H1I8, Q4TT54, Q8TAZ0, Q9H711, Q9H9D6 | Gene names | ASCC2, ASC1P100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 2 (ASC-1 complex subunit p100) (Trip4 complex subunit p100). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.009788 (rank : 146) | θ value | 0.163984 (rank : 53) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
JIP2_MOUSE
|
||||||
| NC score | 0.009754 (rank : 147) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.009360 (rank : 148) | θ value | 8.99809 (rank : 182) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
HCLS1_MOUSE
|
||||||
| NC score | 0.009291 (rank : 149) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.009194 (rank : 150) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.008995 (rank : 151) | θ value | 1.38821 (rank : 88) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
ST5_HUMAN
|
||||||
| NC score | 0.008747 (rank : 152) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
TOPB1_HUMAN
|
||||||
| NC score | 0.008123 (rank : 153) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.007525 (rank : 154) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.007452 (rank : 155) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.007342 (rank : 156) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
SPHK2_HUMAN
|
||||||
| NC score | 0.006731 (rank : 157) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NRA0, Q9BRN1, Q9H0Q2, Q9NWU7 | Gene names | SPHK2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.006677 (rank : 158) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.006643 (rank : 159) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
TUB_HUMAN
|
||||||
| NC score | 0.006271 (rank : 160) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50607, O00293 | Gene names | TUB | |||
|
Domain Architecture |

|
|||||
| Description | Tubby protein homolog. | |||||
|
TYDP1_MOUSE
|
||||||
| NC score | 0.006229 (rank : 161) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ37, Q8VE39 | Gene names | Tdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosyl-DNA phosphodiesterase 1 (EC 3.1.4.-) (Tyr-DNA phosphodiesterase 1). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.006008 (rank : 162) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.005593 (rank : 163) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.005505 (rank : 164) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ANR38_HUMAN
|
||||||
| NC score | 0.005438 (rank : 165) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T7N3, Q6P9A0, Q86T71, Q86VE6, Q8NAX3 | Gene names | ANKRD38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.005417 (rank : 166) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
RAE1_MOUSE
|
||||||
| NC score | 0.005394 (rank : 167) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.005321 (rank : 168) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
POLS_MOUSE
|
||||||
| NC score | 0.005107 (rank : 169) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
CAP2_HUMAN
|
||||||
| NC score | 0.004981 (rank : 170) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | 0.004874 (rank : 171) | θ value | 0.47712 (rank : 74) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
MARCO_MOUSE
|
||||||
| NC score | 0.004430 (rank : 172) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
SNX2_MOUSE
|
||||||
| NC score | 0.003474 (rank : 173) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWK8, Q3UKW3, Q9CQV0 | Gene names | Snx2 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-2. | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.003414 (rank : 174) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
BRAC_HUMAN
|
||||||
| NC score | 0.002207 (rank : 175) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15178 | Gene names | T | |||
|
Domain Architecture |

|
|||||
| Description | Brachyury protein (T protein). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.001783 (rank : 176) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
PAK4_HUMAN
|
||||||
| NC score | 0.000582 (rank : 177) | θ value | 0.365318 (rank : 65) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O96013, Q8N4E1, Q8NCH5, Q8NDE3, Q9BU33, Q9ULS8 | Gene names | PAK4, KIAA1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
CADH6_HUMAN
|
||||||
| NC score | 0.000396 (rank : 178) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55285, Q9BWS0 | Gene names | CDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
CADH6_MOUSE
|
||||||
| NC score | 0.000295 (rank : 179) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97326, P70393 | Gene names | Cdh6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | -0.000339 (rank : 180) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | -0.000345 (rank : 181) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | -0.000396 (rank : 182) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CE152_HUMAN
|
||||||
| NC score | -0.000622 (rank : 183) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
PITX1_HUMAN
|
||||||
| NC score | -0.001051 (rank : 184) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78337, O14677, O60425, Q9BTI5 | Gene names | PITX1, BFT, PTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 1 (Hindlimb expressed homeobox protein backfoot). | |||||
|
GAK_MOUSE
|
||||||
| NC score | -0.002034 (rank : 185) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
M3KL4_HUMAN
|
||||||
| NC score | -0.003083 (rank : 186) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5TCX8, Q5TCX7, Q5TCX9, Q8WWN1, Q8WWN2, Q96JM1 | Gene names | MLK4, KIAA1804 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase (EC 2.7.11.25) (Mixed lineage kinase 4). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | -0.003391 (rank : 187) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
LRRK1_MOUSE
|
||||||
| NC score | -0.003985 (rank : 188) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHC2, Q3U476, Q66JQ4, Q6GQR9, Q6NZF5, Q6ZPI4, Q8BKP3, Q8BU93, Q8BUY0, Q8BVV2, Q8R085 | Gene names | Lrrk1, Kiaa1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||